Installation Steps: BioTuring BioStudio
¶Introduction ¶
Software to tackle biomedical challenges:
Colaboratory , or BioStudio for short, is a product from Bioturing.BioStudio has a variety of features and pre-built notebooks , for user to download and use. We are providing all types of user friendly tools that helps, users to post their data and analyze the reports. Our product can be used to write and execute arbitrary python, R code, Golang, Julia, RStudio, VS Code and many more through the browser, and is especially well suited to data analysis and education. More technically, BioStudio is a hosted Jupyter notebook service that requires no setup to use. Users can build their own notebook based on their requirements.
We are keep adding new features and functionalities to BioStudio.
System Requirements ¶
Before installing the BioStudio, some pre-installation steps are required:
| Basic recommendation | Optional | |
|---|---|---|
| CPU | 16 core | This is basic requirement to start BioStudio and based on requirement, Resources as well as machine can be added. |
| RAM | 64 GB Minimum | As above |
| HDD | / partition can be 100 GB | As above. |
| Data Volume : 1TB | As above. | |
| OS | Any OS. Ubuntu 20.04 and above. | BioStudio is more supportive with Linux OS. For better performance linux OS is recommended. |
| AWS Instance | Support any type of instance type. Depend on needs | AWS g5.4xlarge in case using GPU. |
| Platform | Docker / Kubernetes |
| Note | |
|---|---|
| In case we want to use GPU based Notebooks with BioStudio. We need to install NVIDIA and below would be a requirement. | |
| The system has one or multiple NVIDIA GPU(s) (at least 16 GB memory per GPU) - with Turing architecture or above. | |
| BioStudio supports any Linux OS. We are recommending Ubuntu 20.04 or above. | |
| SSL can be configured later also. | |
| Please contact support@bioturing.com to get the token for your company. | |
| Security | |
| The BioStudio platform uses HTTPS protocol to securely communicate over the network. | |
| All of the users need to be authenticated using a BioTuring account or the company’s SSO to access the platform. | |
| We highly recommend setting up a private VPC network for IP restriction. | |
| The data stays behind the company firewall. | |
| The BioStudio platform does not track any usage logs. | |
| Data visibility | |
| Data can be uploaded to Personal Workspace or Data Sharing group. | |
| In the Personal Workspace, only the owner can able to see and manipulate the data she/he uploaded. | |
| In the Data Sharing group, only people in the group can able to see the data. | |
| In the Data Sharing group, only people with sufficient permissions can able to manipulate the data. |
Network requirements ¶
| Domain | Explain |
|---|---|
| *.bioturing.com | We need to retrieve data from the BioTuring ecosystem server. |
| *.anaconda.org | We need to retrieve packages from the Anaconda server. |
| *repo.anaconda.com | We need to retrieve packages from the Anaconda repo server. |
| Amazon S3 | We need to retrieve resources from the BioTuring ecosystem server. |
| github.com | We need to retrieve packages from the Github. |
| cdn-eu-west-1.s3.eu-west-1.amazonaws.com (euro -west1) | We need to retrieve packages from the CDN. |
| s3.us-west-2.amazonaws.com/cdn.bioturing.com (us -west2) | We need to retrieve packages from the CDN. |
| cdn.bioturing.com (us -west2) | We need to retrieve packages from the CDN. |
| talk2dataupdate.s3.us-west-2.amazonaws.com | We need to retrieve packages from here. |
| openapi.bioturing.com | We need to retrieve packages from here. |
| portal.bioturing.com | We need to retrieve packages from here. |
| update.bioturing.com | We need to retrieve packages from here. |
| talk2data.bioturing.com | We need to retrieve packages from here. |
| ml.bioturing.com | We need to retrieve packages from here. |
| authenticate.bioturing.com | We need to retrieve packages from here. |
| colab.bioturing.com | We need to retrieve packages from here. |
| cdn.bioturing.com | We need to retrieve packages from here. |
| studio.bioturing.com | We need to retrieve packages from here. |
👉 Please contact 📧 support@bioturing.com to get the token for your company.
🔔 Note: Our software can be installed with minimum capacity of instance / server. The ideal system that we recommend for most companies is AWS c5a.8xlarge for CPU based. Instance can be chosen based on requirement. If the notebook is based on GPU, we can select GPU based instance. Our Product is containerized based applications. Kindly select the machine based on your requirements. BioStudio can be run on Docker using Docker engine and Kubernetes.
✨ Important note ✨: We can install BBrowserX talk2data (Bioturing ecosystem) 🔗 BBrowserX and BioStudio on the same server. Installation process is the same to run our installation script for BioStudio. Step by step Instructions to install BioStudio are given below. Only changes on domain name for BioStudio. Kindly do not use BBrowserX talk2data domain name during this installation, In case you are planing to install BBrowserX and BioStudio on same machine.
💫 BioStudio token can be updated later, after installation.
💫 Contact point to get BioStudio token is 📧 support@bioturing.com
💫 SSL certificate can be installed later, If BioStudio planned to install on separate instance.
User can access BioStudio application using two ways, In case installed BBrowserx and BioStudio on the same machine.
1️⃣ Browse the BioStudio application https://<BioStudio Domain name> , If you have SSL certificate for BioStudio domain and configured with Nginx else use http protocol.
2️⃣ Use BioStudio Desktop application - using IP address.
Bioturing Launcher 🔗 Desktop Application
🔔If BioStudio will install on separate server. BioStudio can be accessed using any browser by IP address or Domain name.
Download and Install ¶
Note: We suggest starting from scratch to avoid package/driver conflicts. For Tag naming conversion, kindly select based on your architecture.
New Server Set up on AWS. ¶
🔶 Login to AWS console with admin user account to launch an EC2 instance.
Note: It’s up to the client. How they are going to manage infrastructure, Load, Network, Access and traffic …etc.
Create a VPC ¶
🔶 Search VPC on the search box.
🔶 Select the appropriate region for VPC.
🔶 Click on *Create VPC.
🔶 Fill-out Name tag , IPv4 CIDR block details.
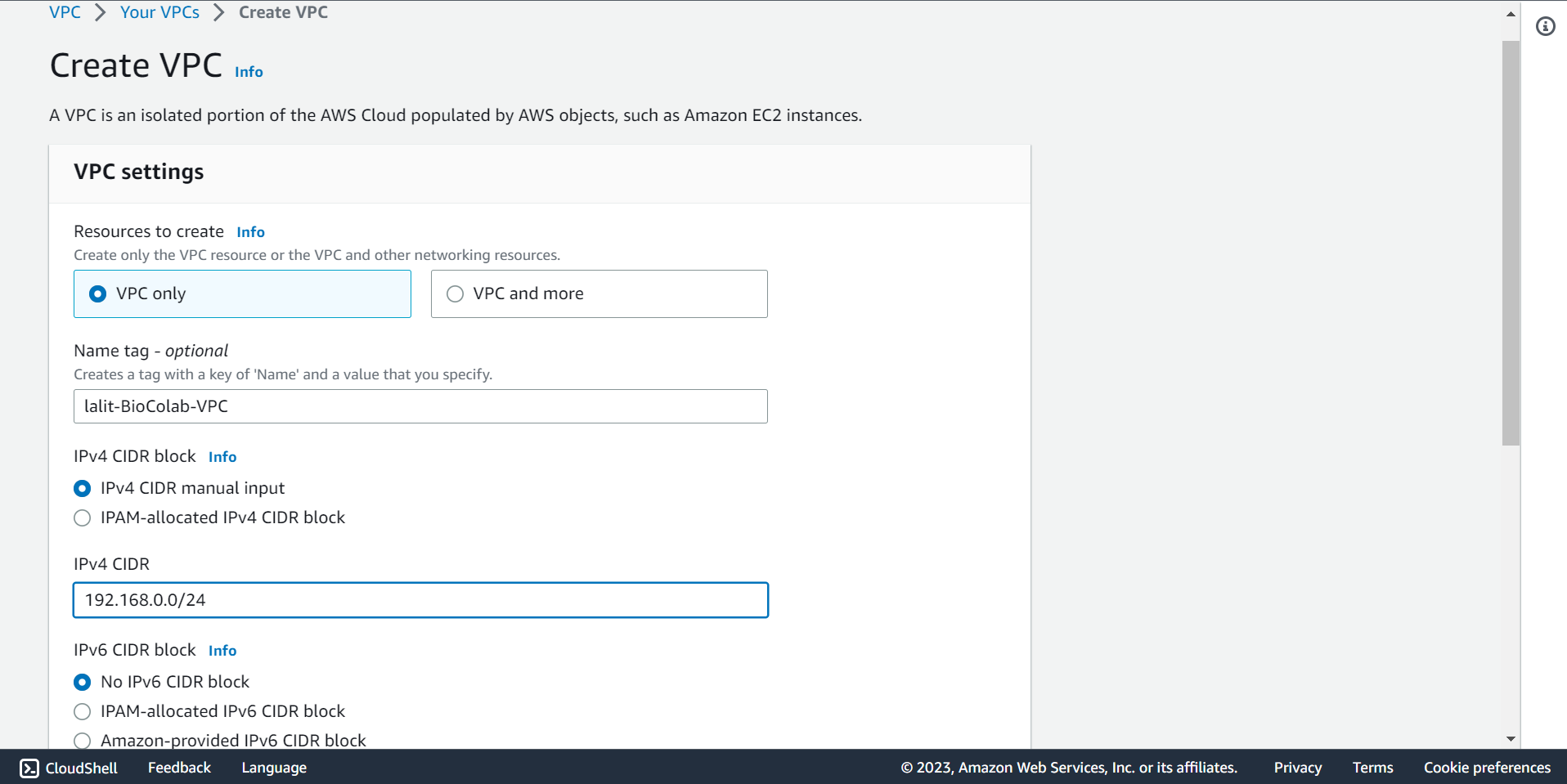
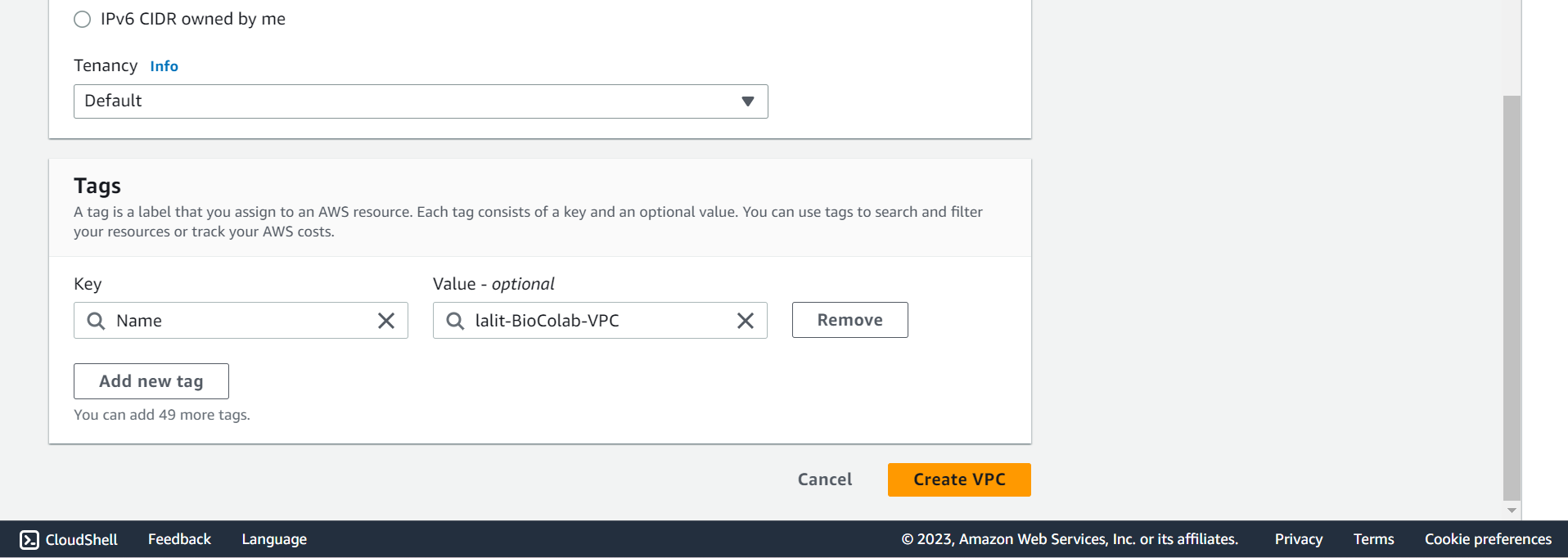
🔶 Mentioned Tags and select Create VPC push button.
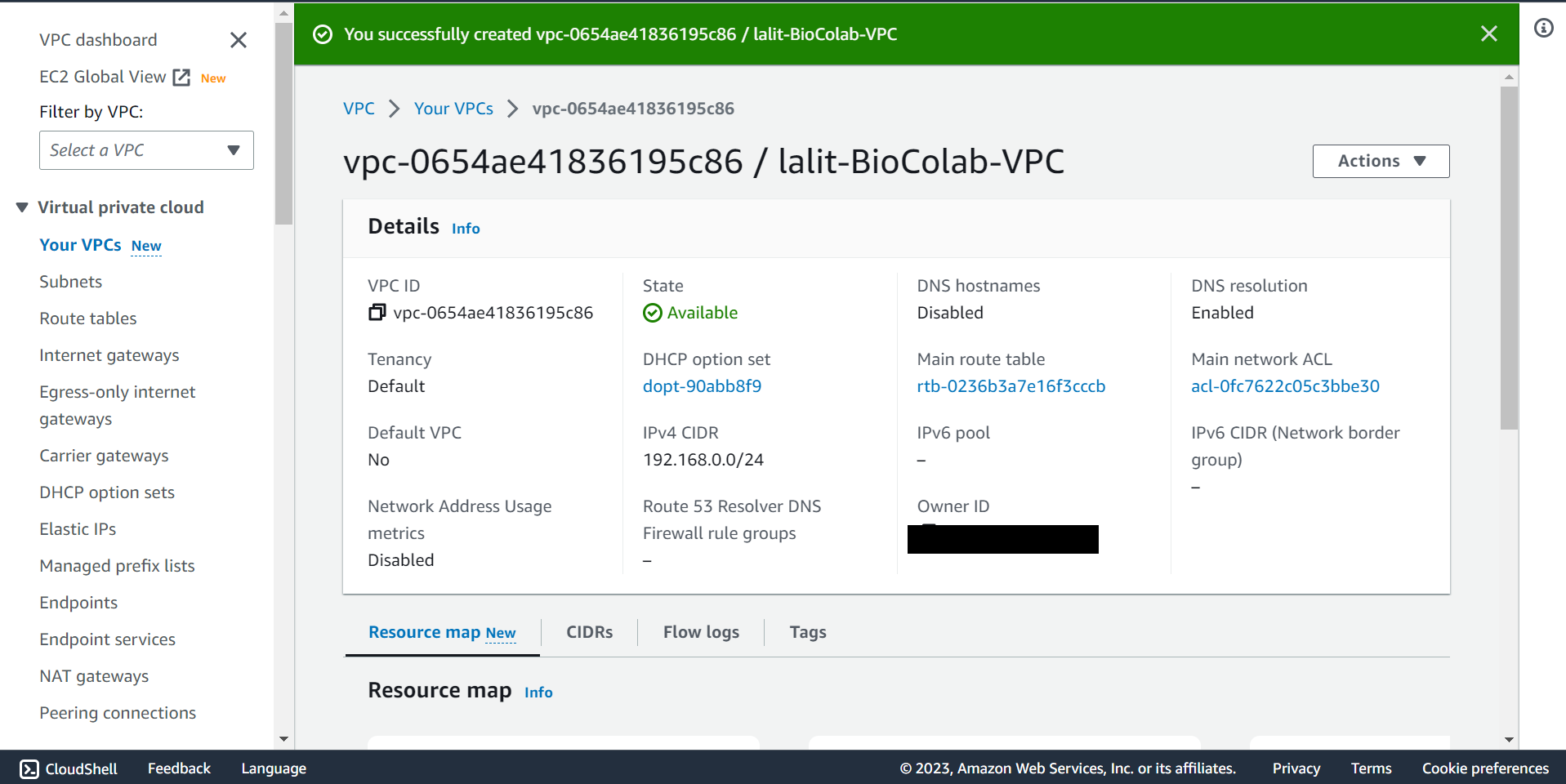
🔶 Verify VPC creation successfully completed.
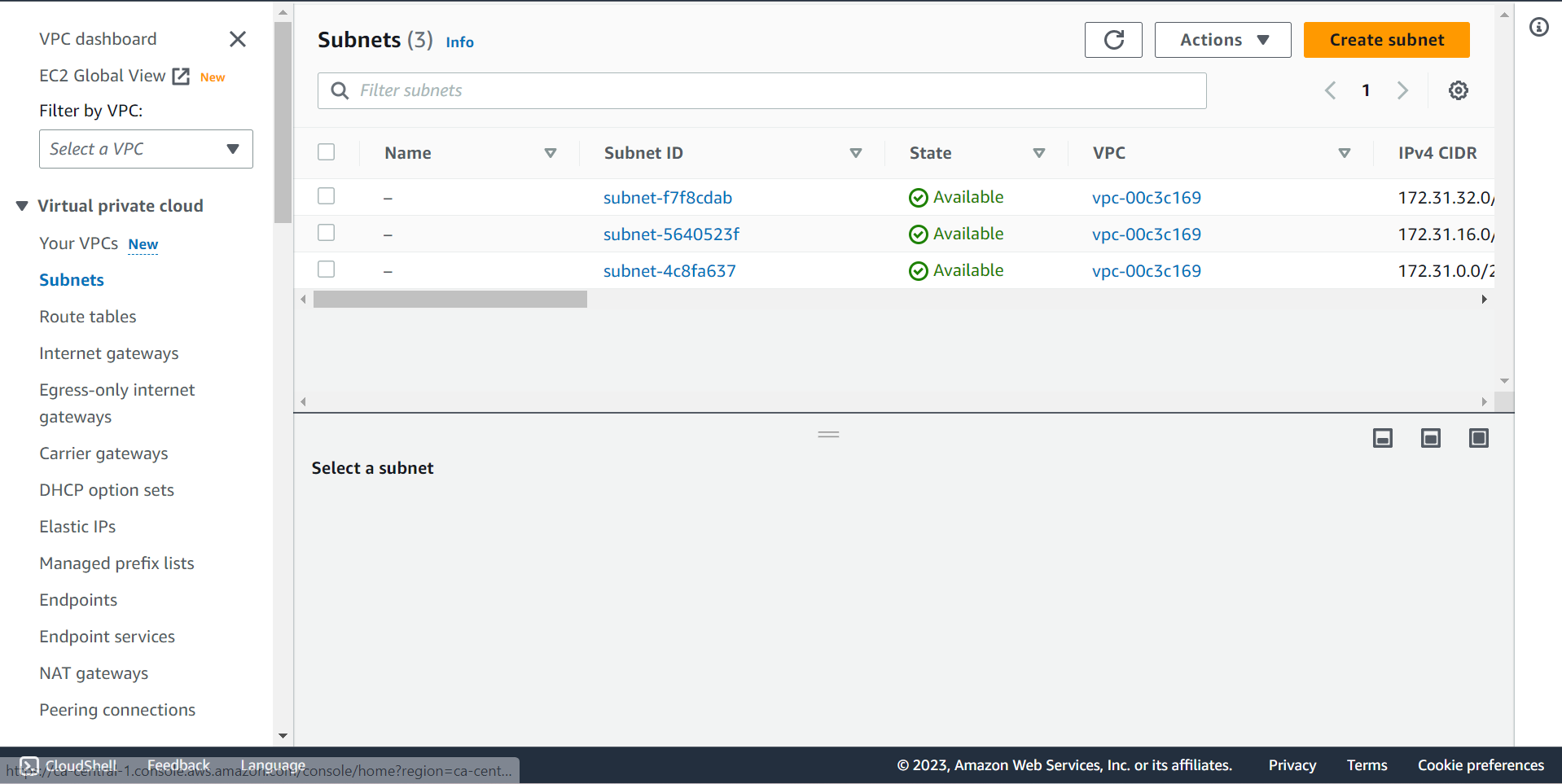
Create a Subnet ¶
🔶 Time to create Subnet. Click on Create subnet.
🔶 Select VPC ID and type Subnet name.
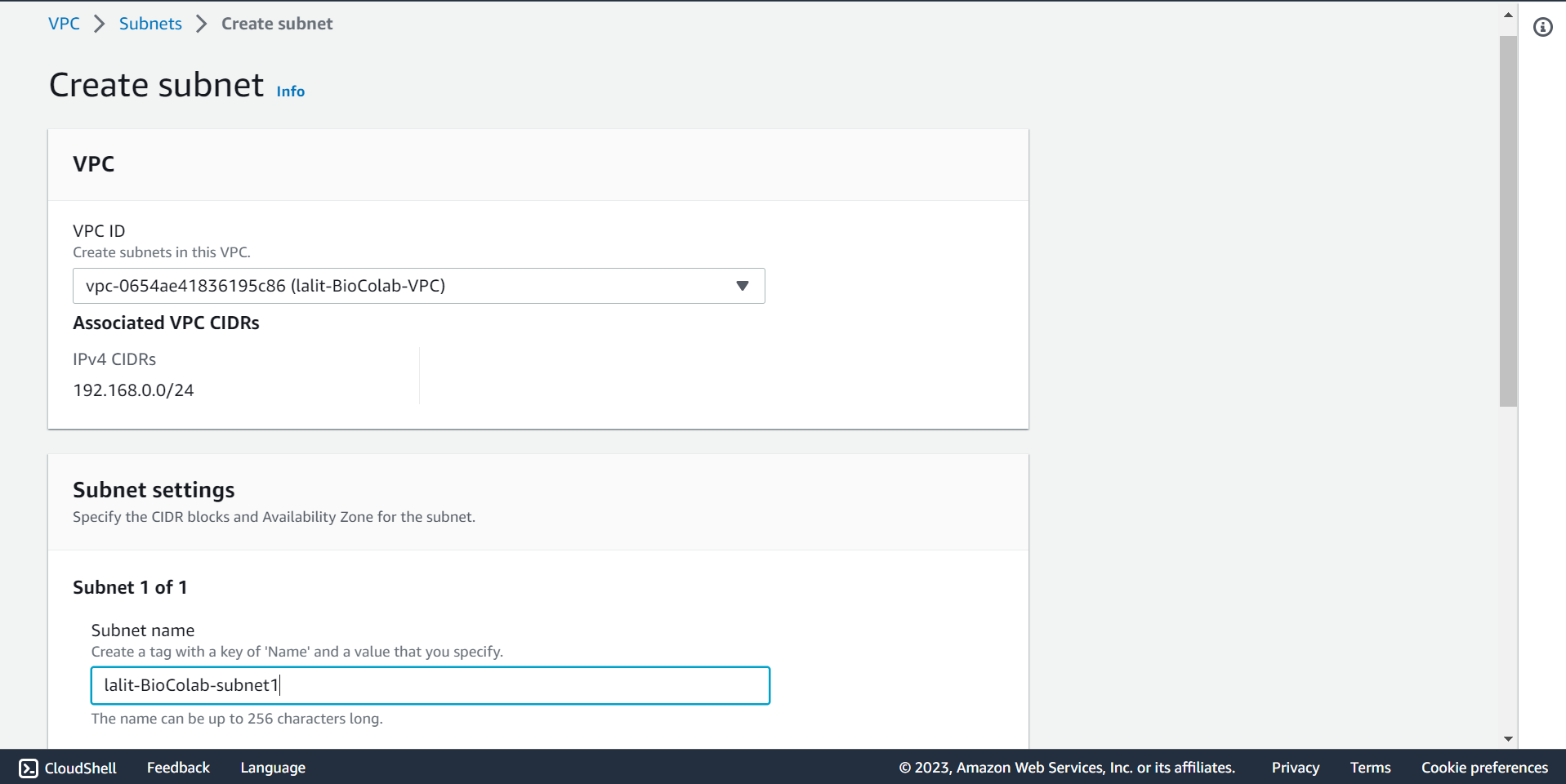
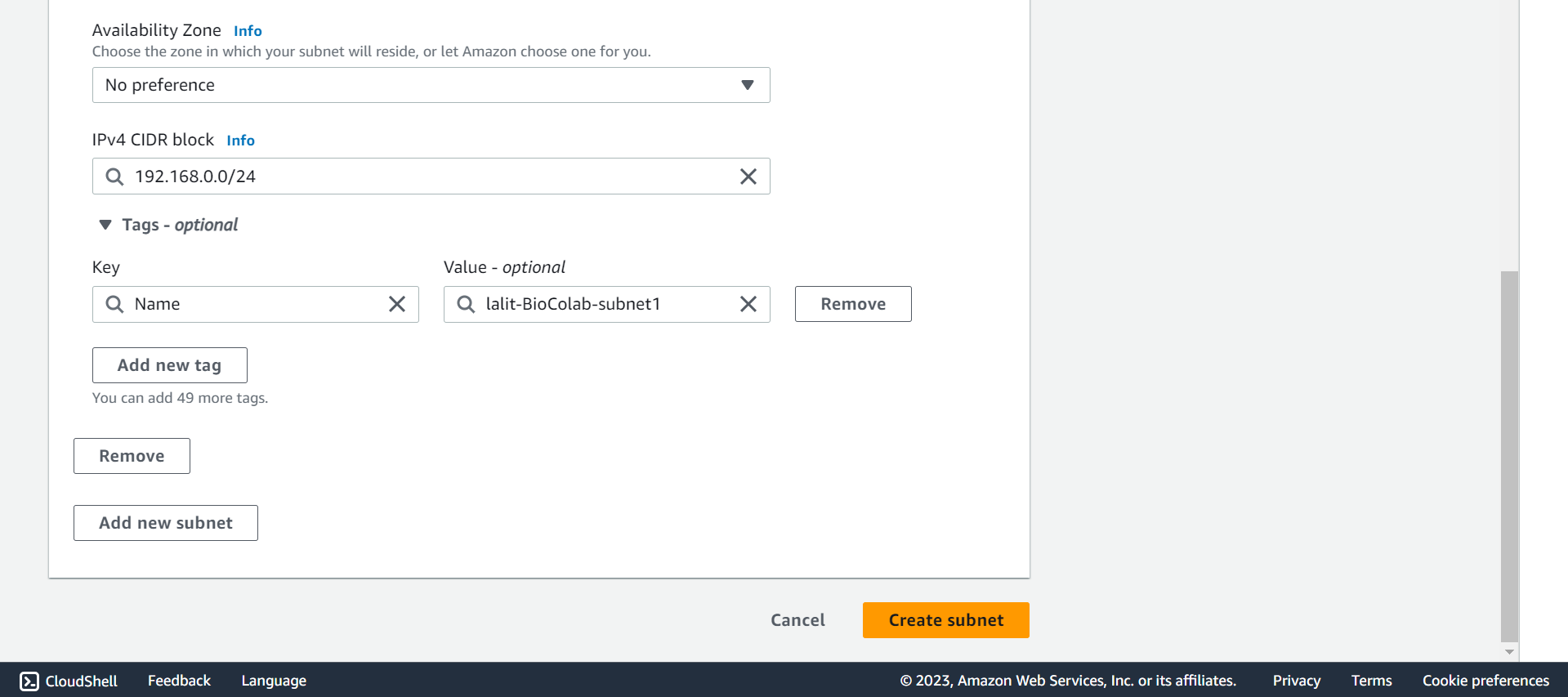
🔶 Fill CIDR block, type Tag Key and Value then Click on Create subnet.
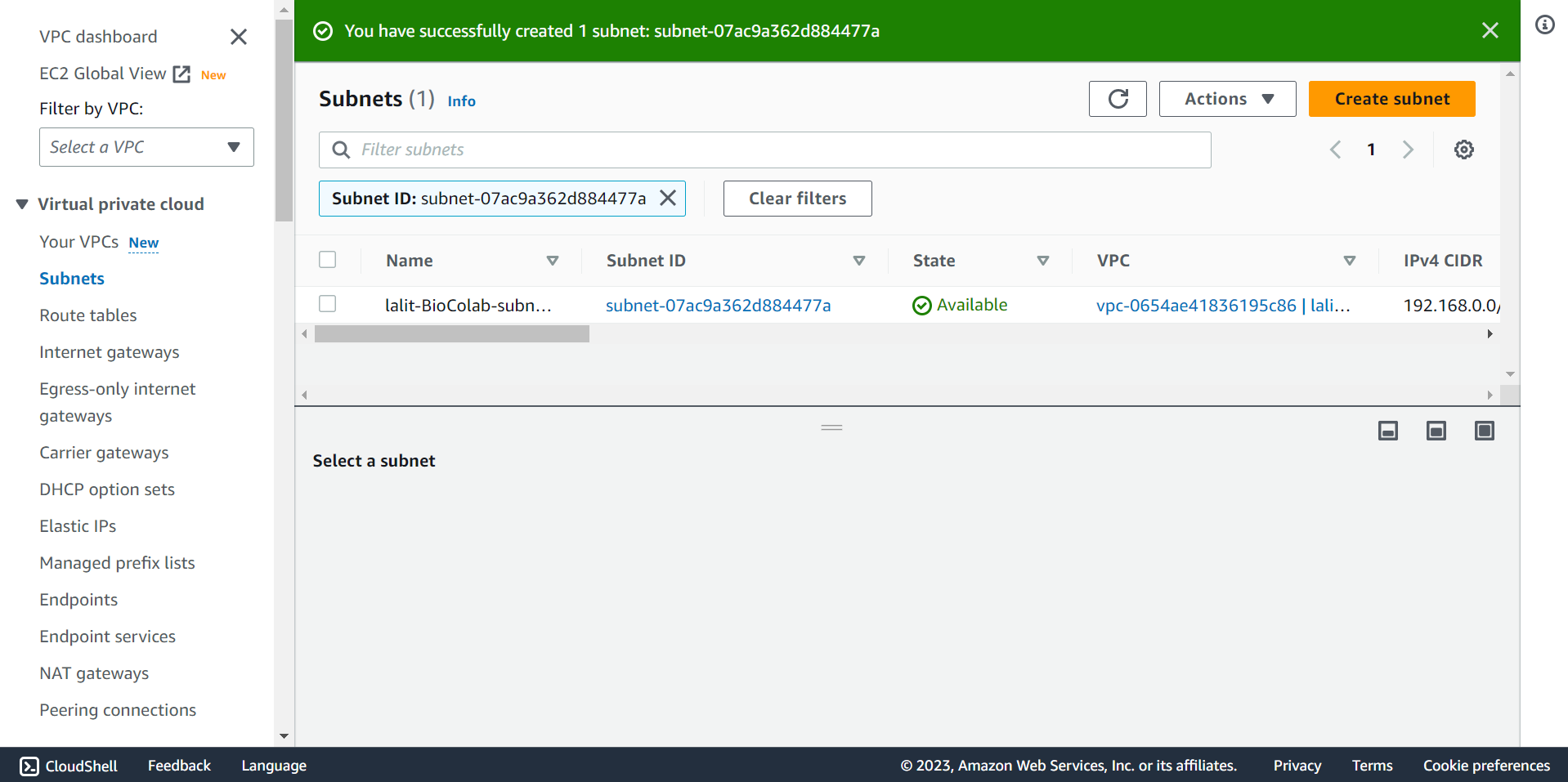
🔶 Subnet created successfully.
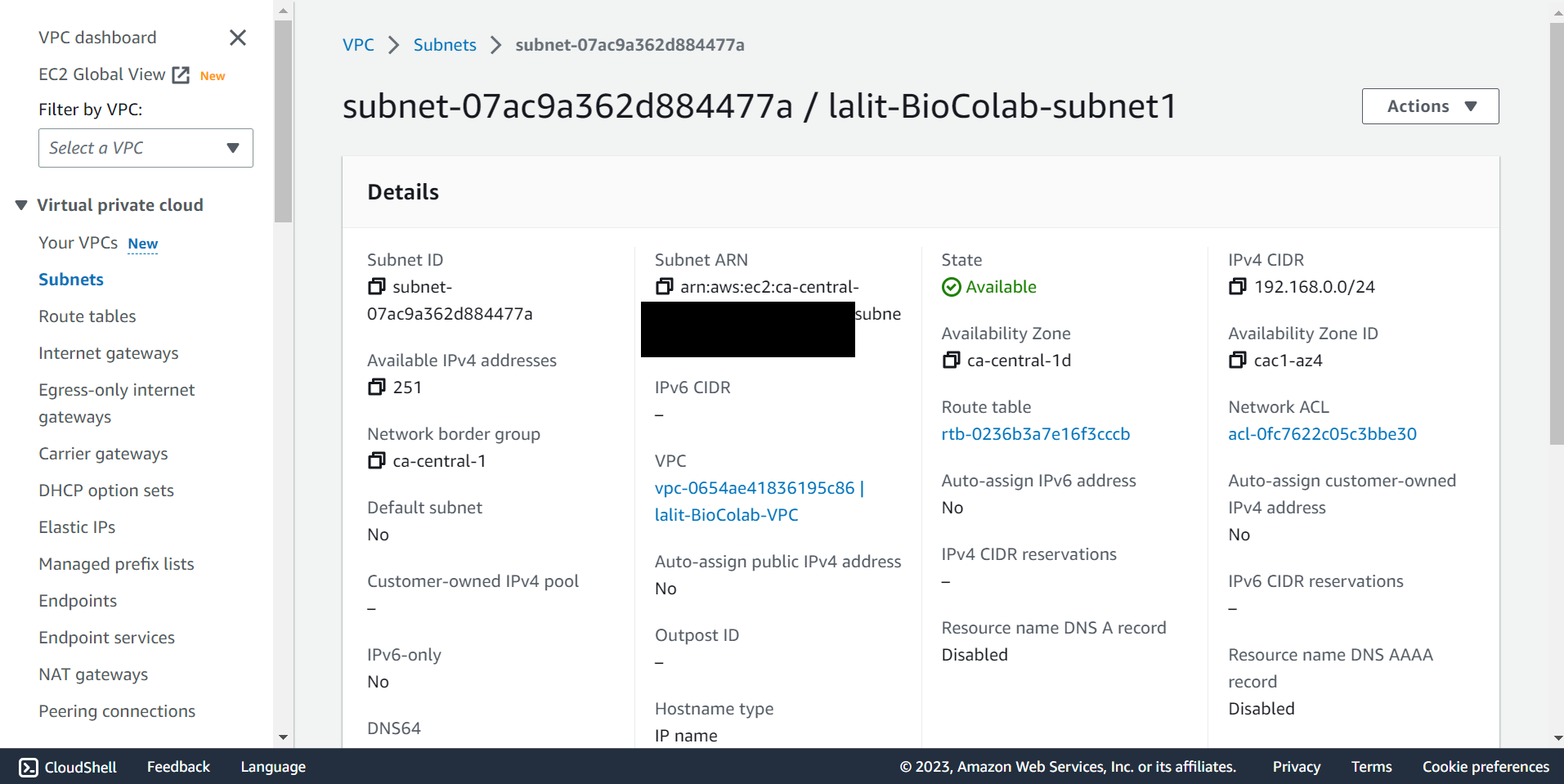
🔶 Check Subnet detail.
Create Internet Gateway ¶
Note: As noticed. During Subnet creation 🔃 Router table was automatically created.
⭕ Time to create Internet Gateway.
🔶 Router Table.
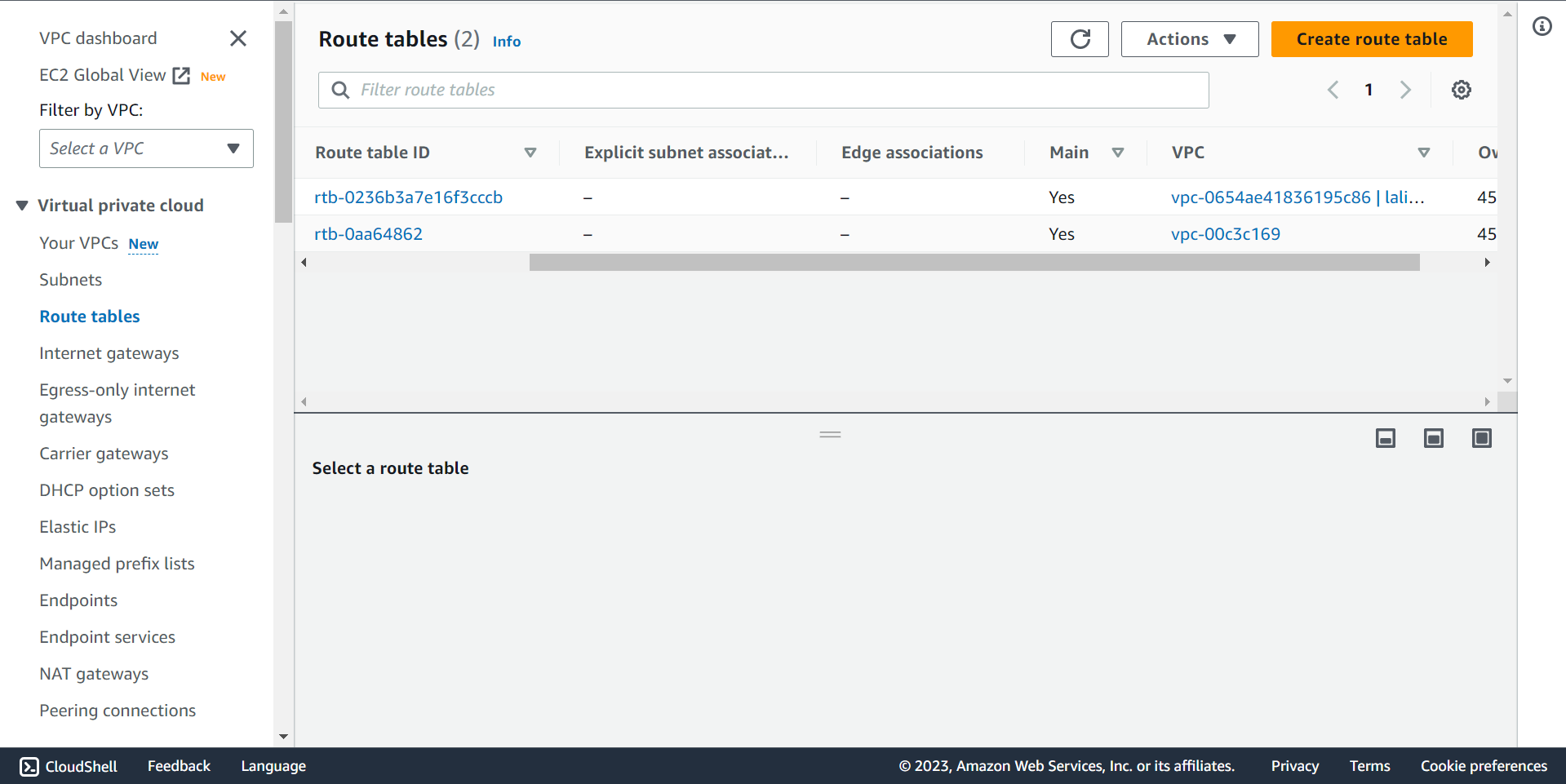
🔶 Select Internet gateway and Click on Create internet gateway.
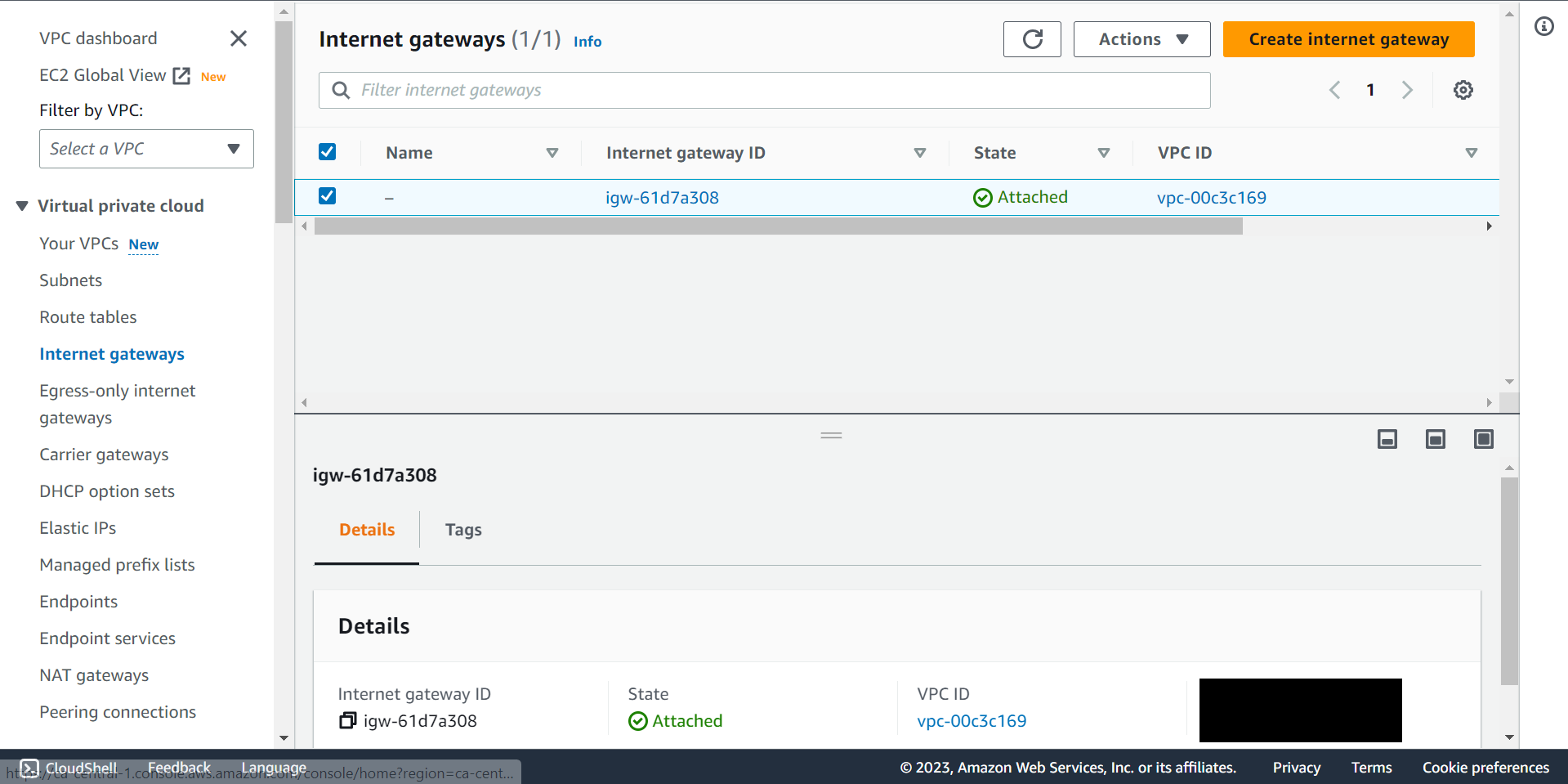
🔶 Fill Name Tag, Key and Value then Click Create internet gateway.
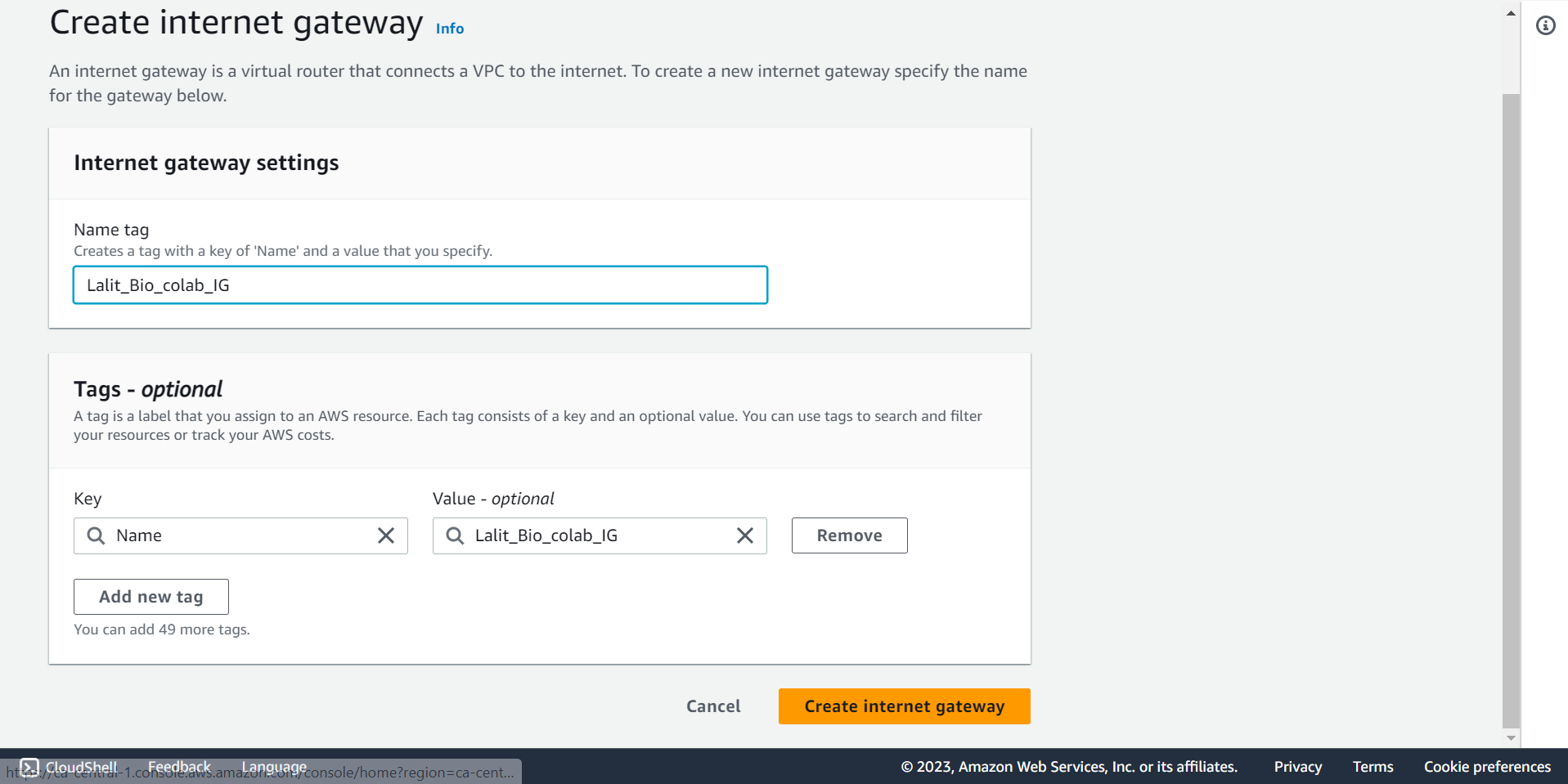
🔶 Internet gateway created successfully.
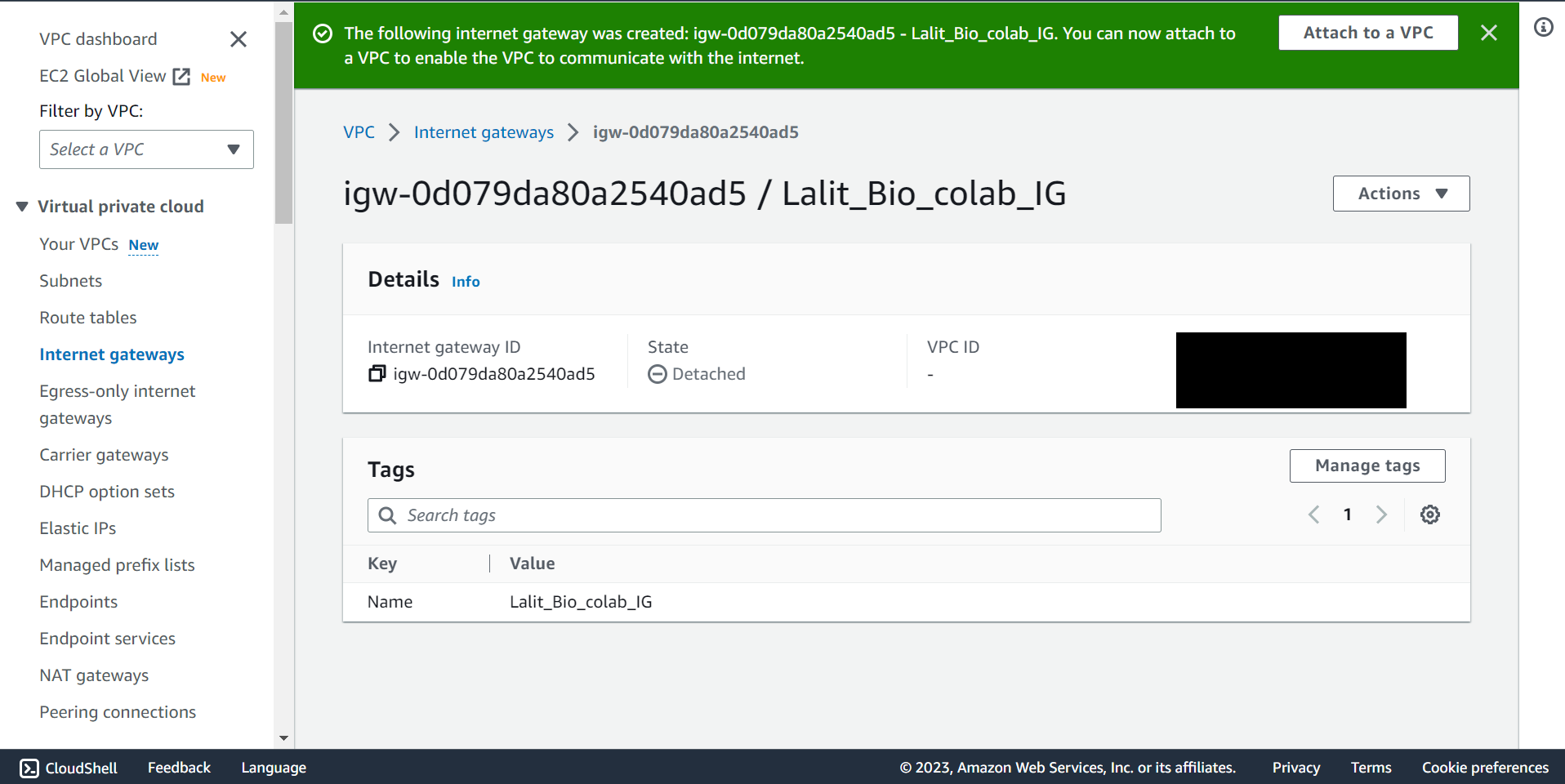
Internet Gateway attached to VPC. ¶
🔶 In Internet gateway Click on Action and select Attached to VPC.
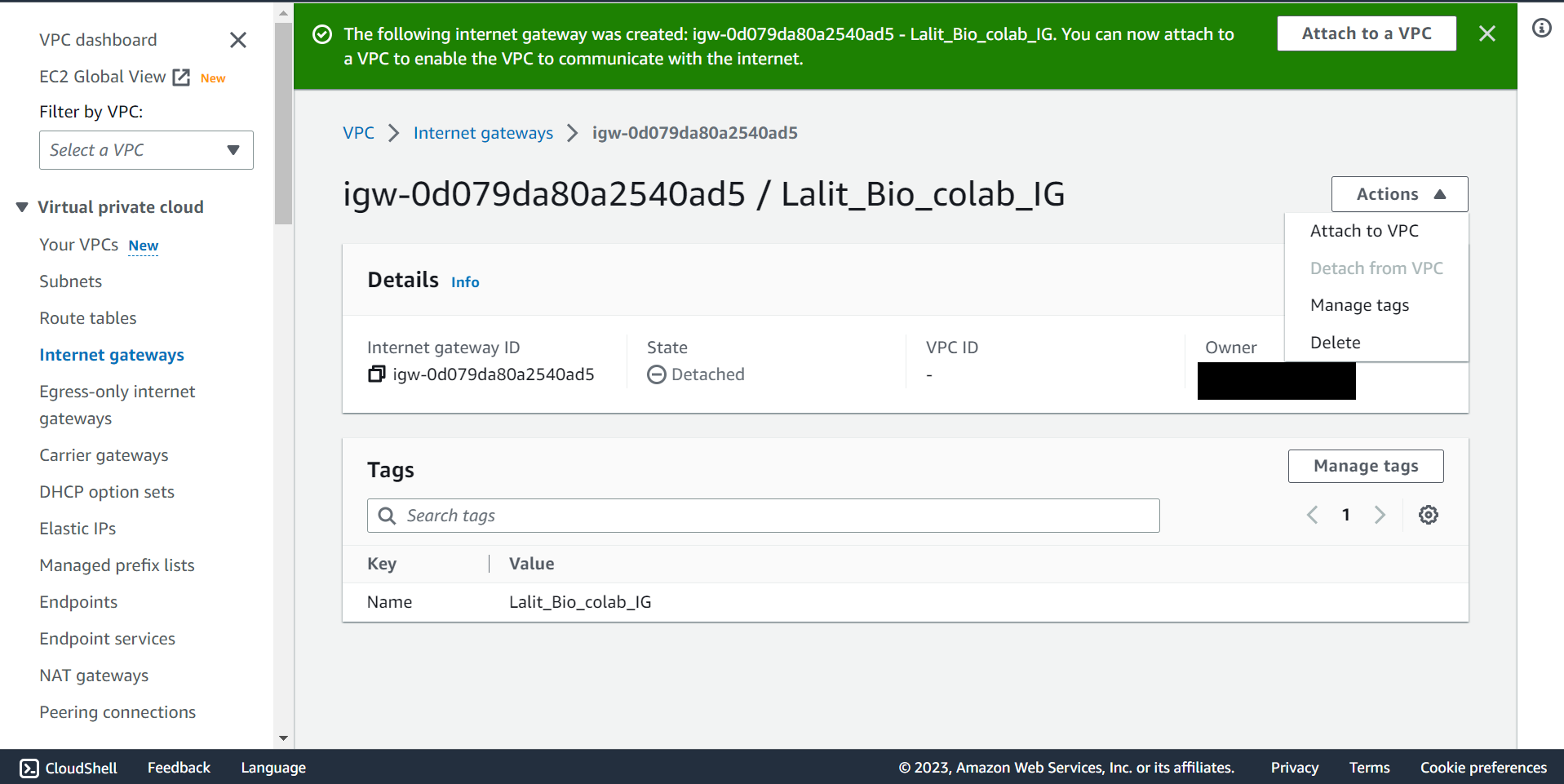
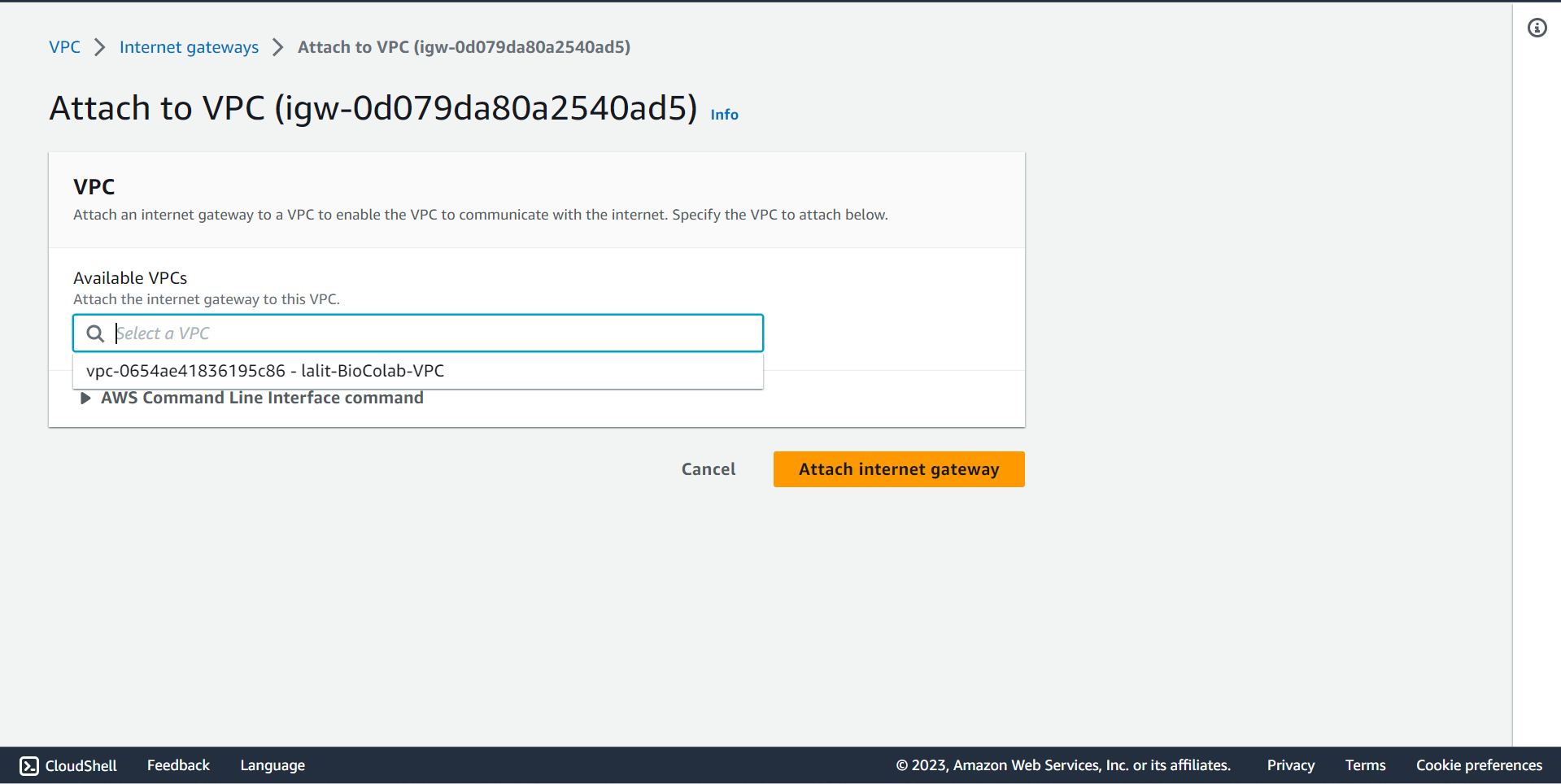
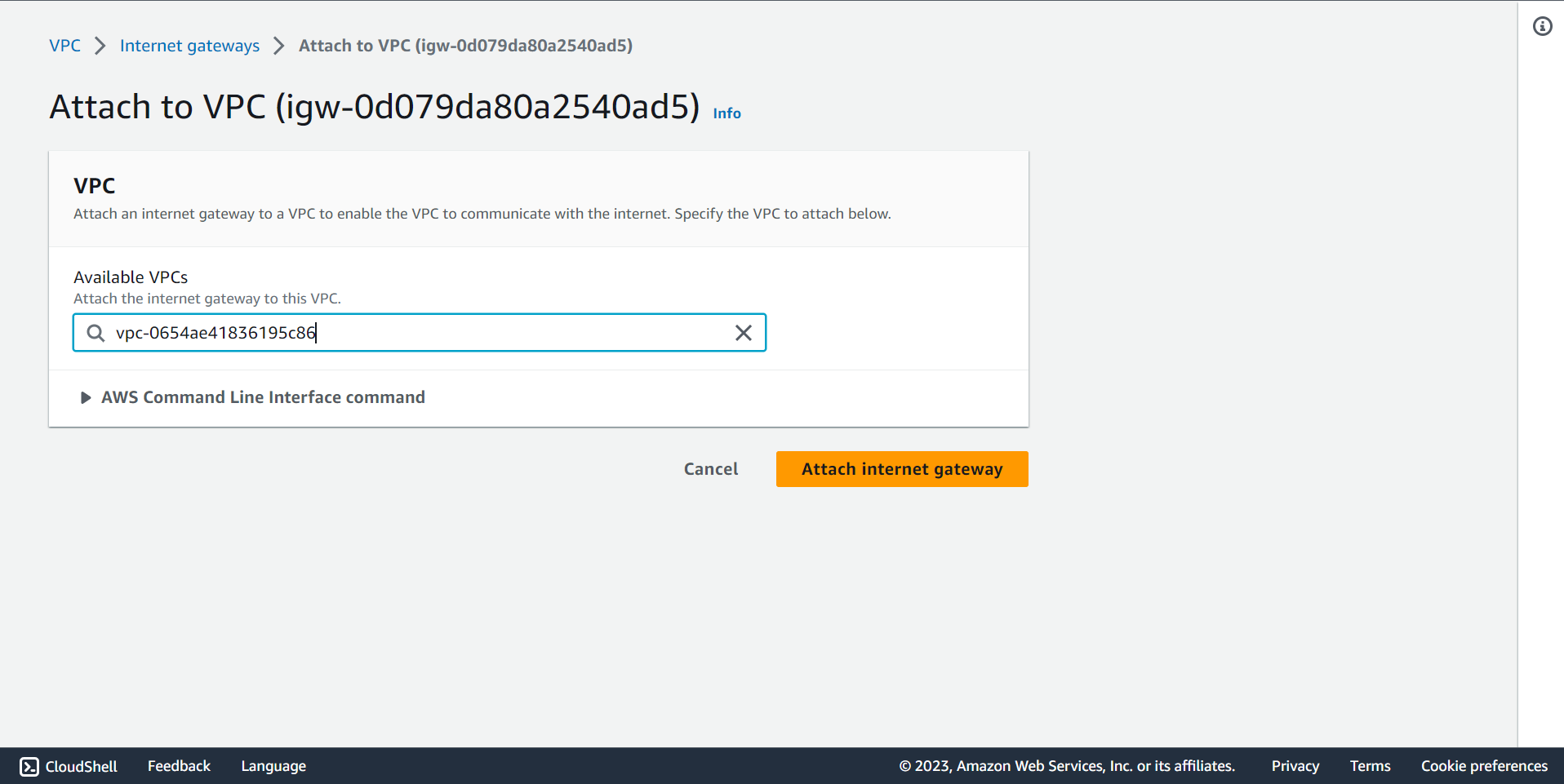
🔶 Internet Gateway attached to VPC completed.
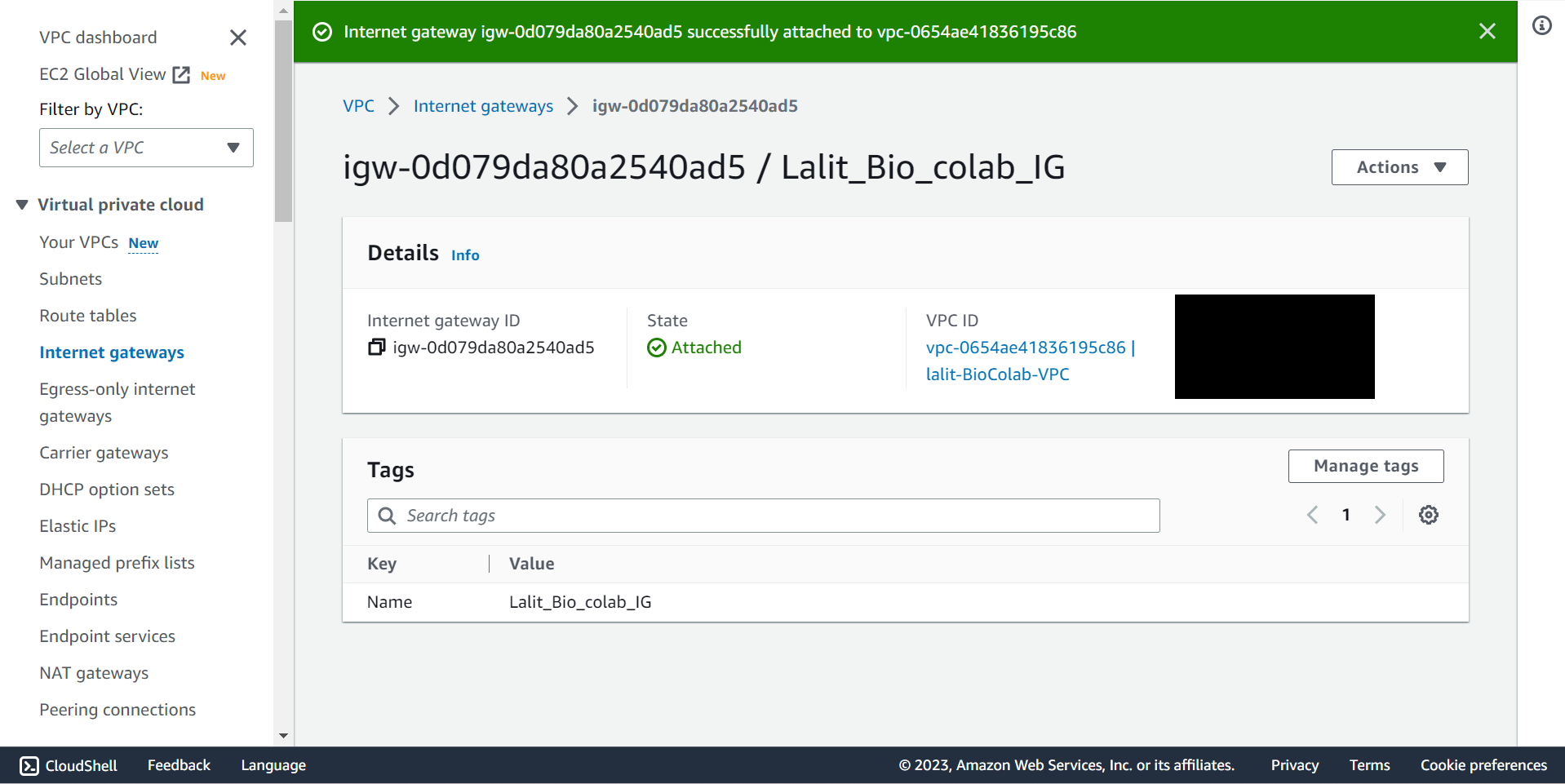
🔶 Now Update Route Table for Subnet association and add Route to use Internet gateway to router table.
🔶 Update Route Table for Subnet association.
Subnet association to Router Table. ¶
🔶 Select Subnet associations from Route tables ▶️ Second tab.
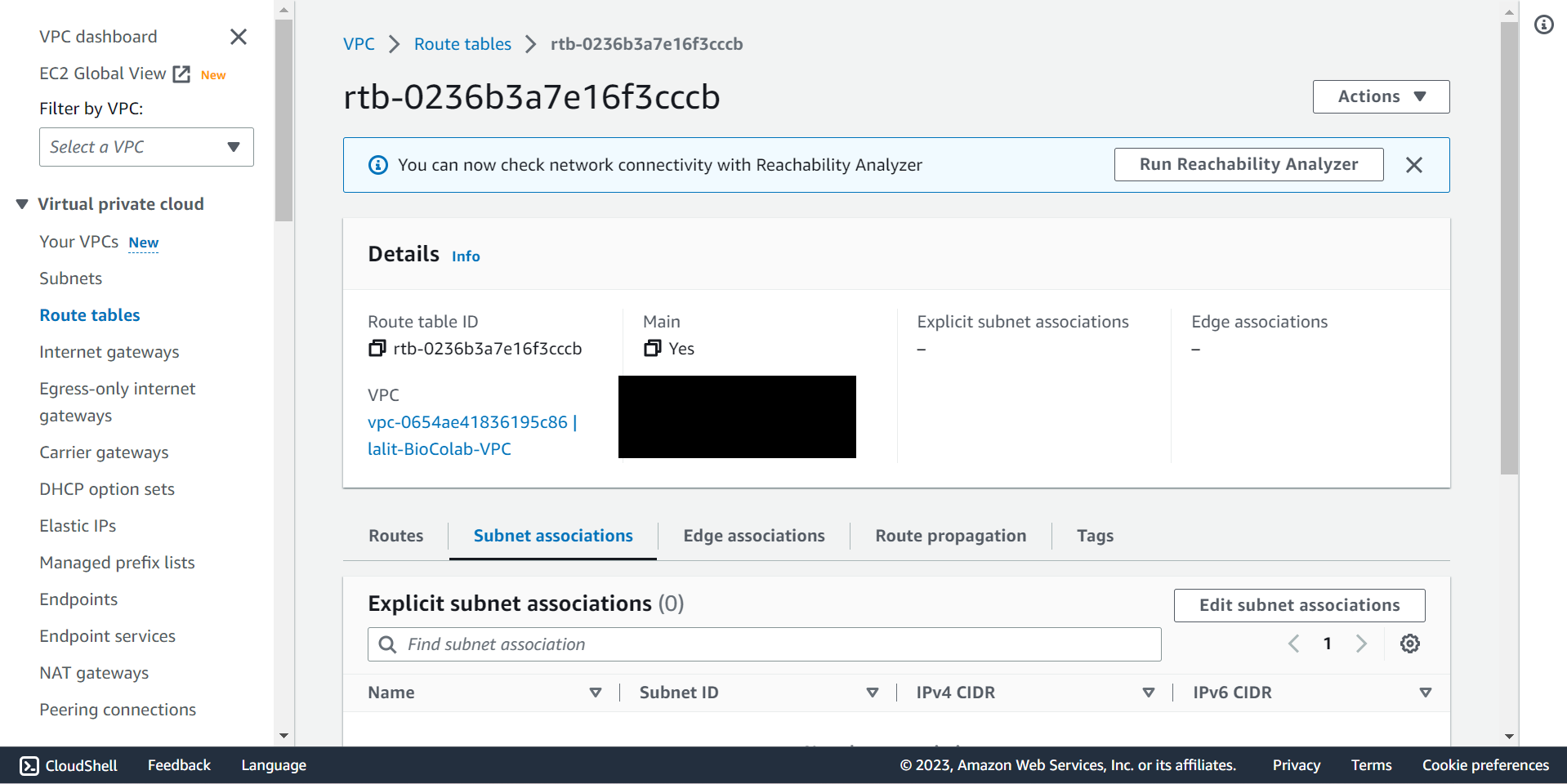
🔶 Click on Edit subnet associations and select Subnet name, which you would like to associate then Click on Save associations button.
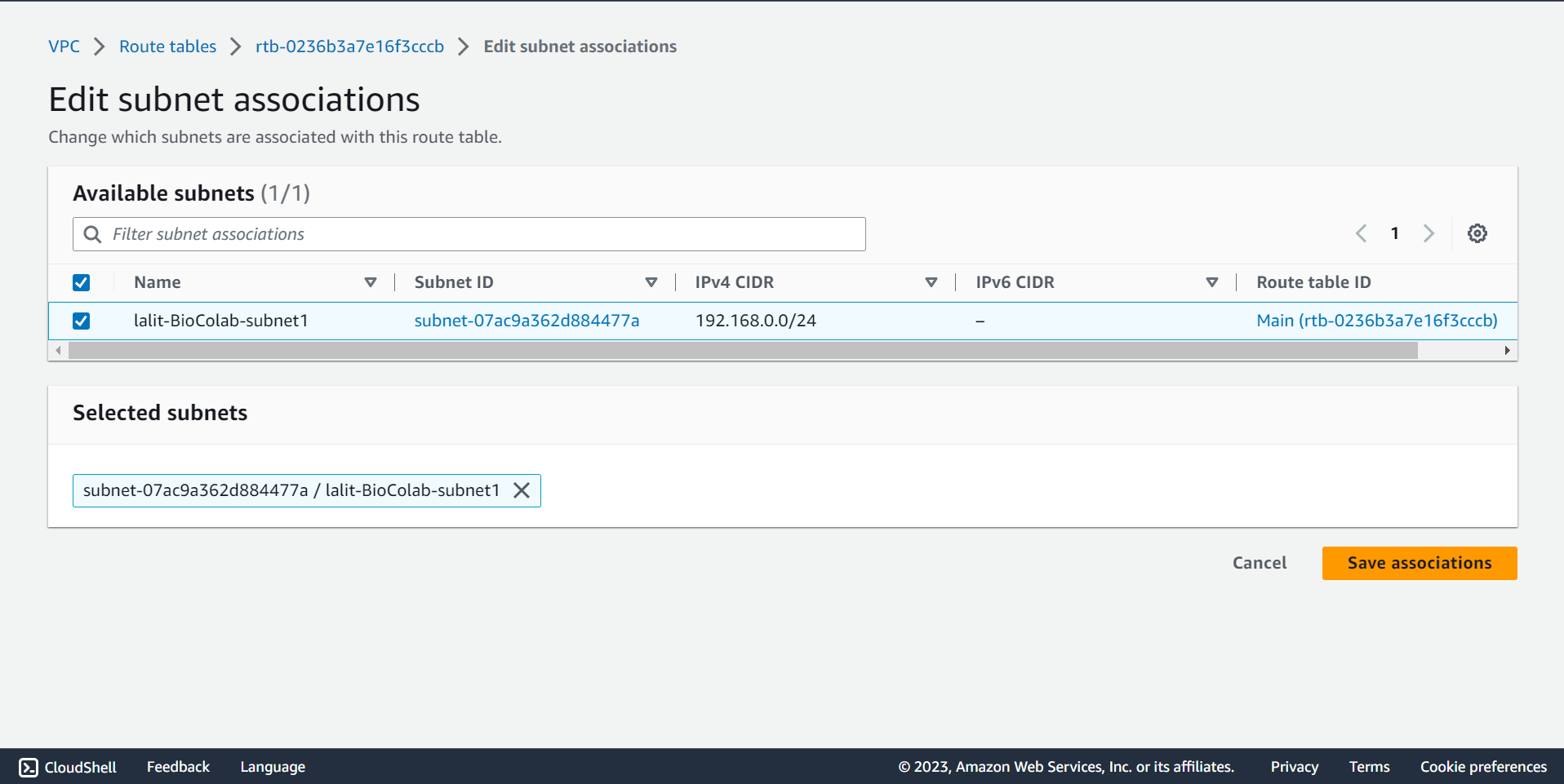
🔶 Subnet association completed.
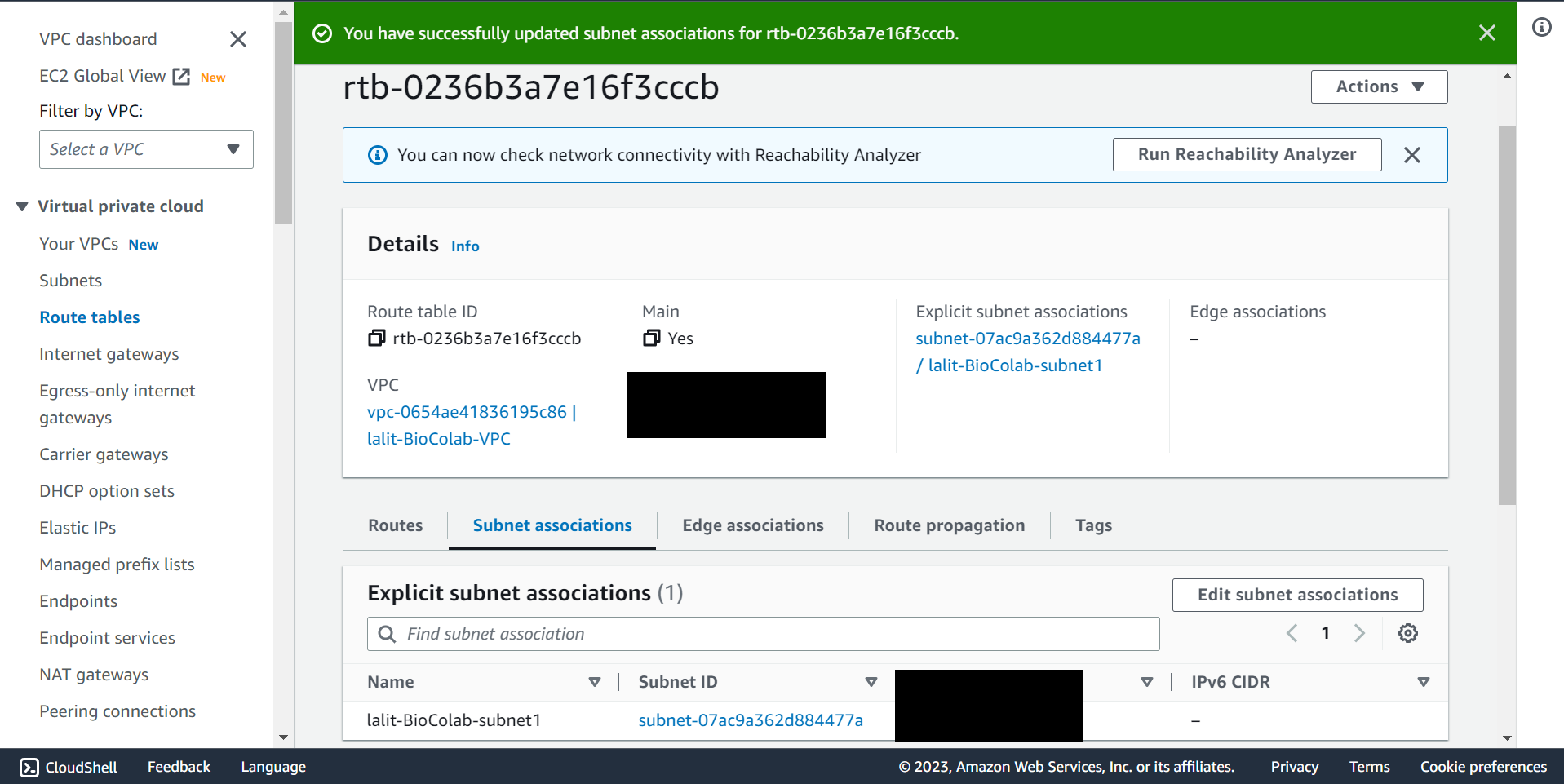
Update Router table to use Internet gateway. ¶
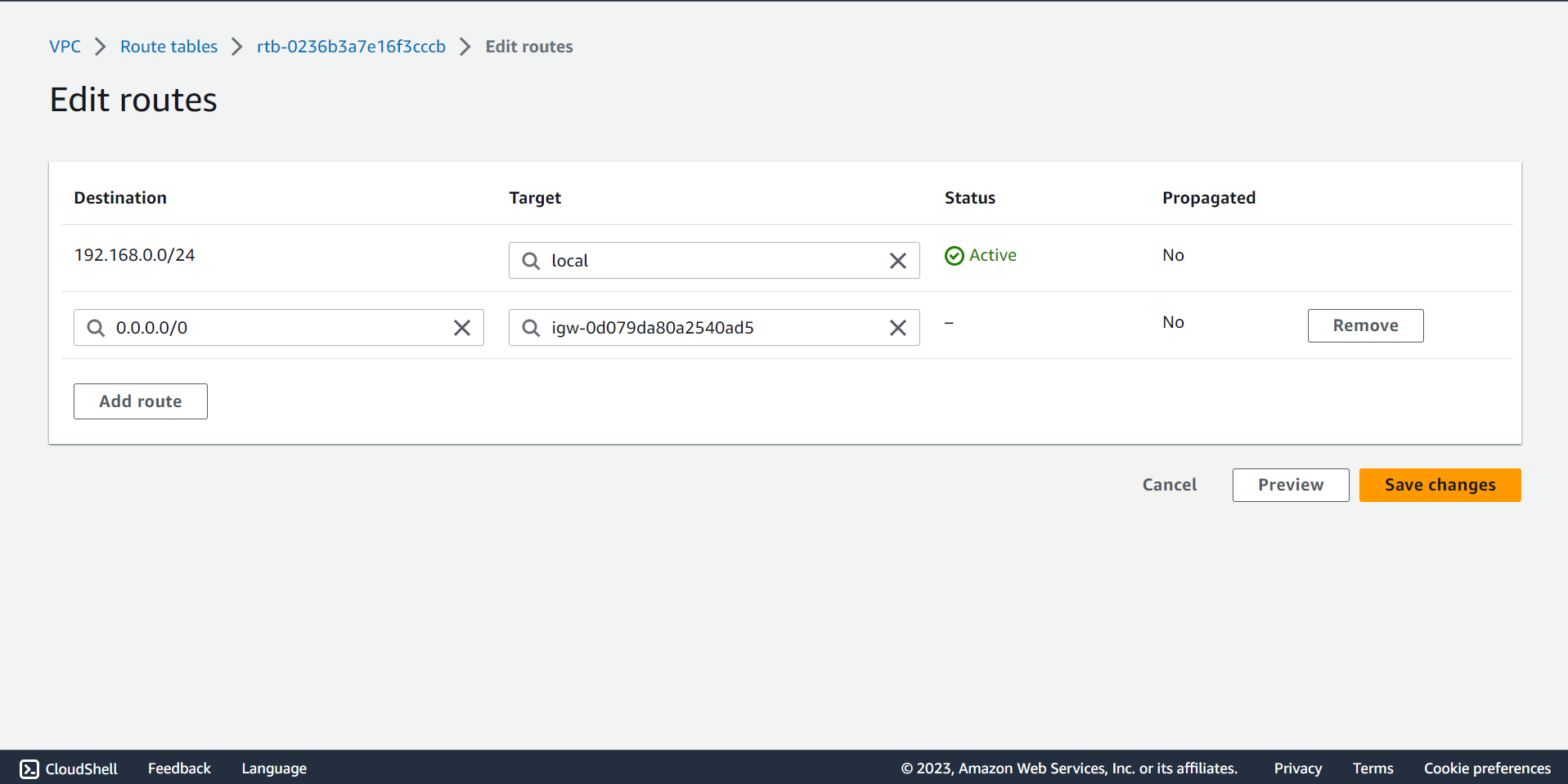
🔶 Select Routes from Route tables ▶️ First tab.
🔶 Click on Edit routes.
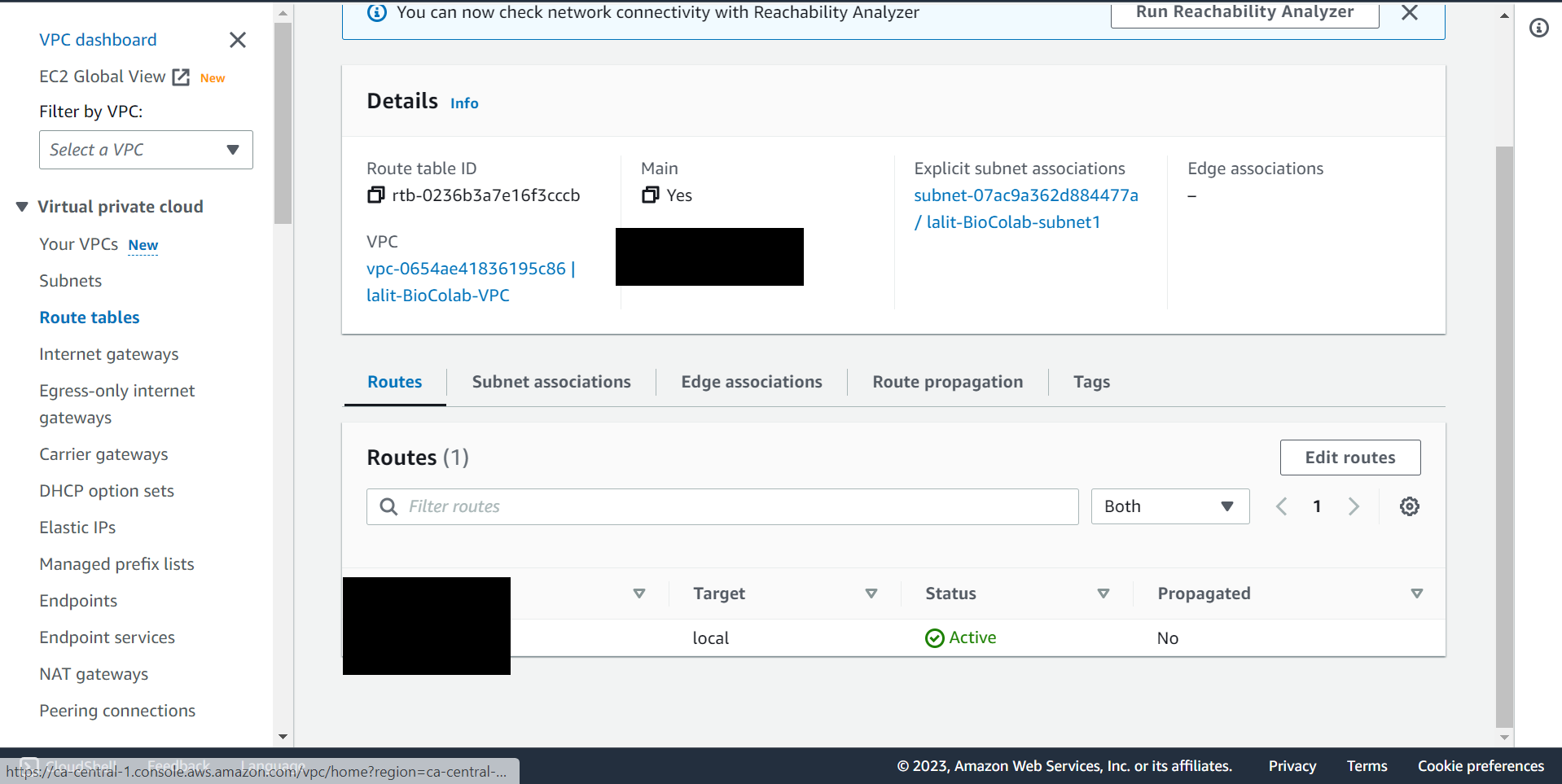
🔶 Fill the value 0.0.0.0/0 and select Internet gateway name as showing on below screenshot.
🔶 Click on Save change.
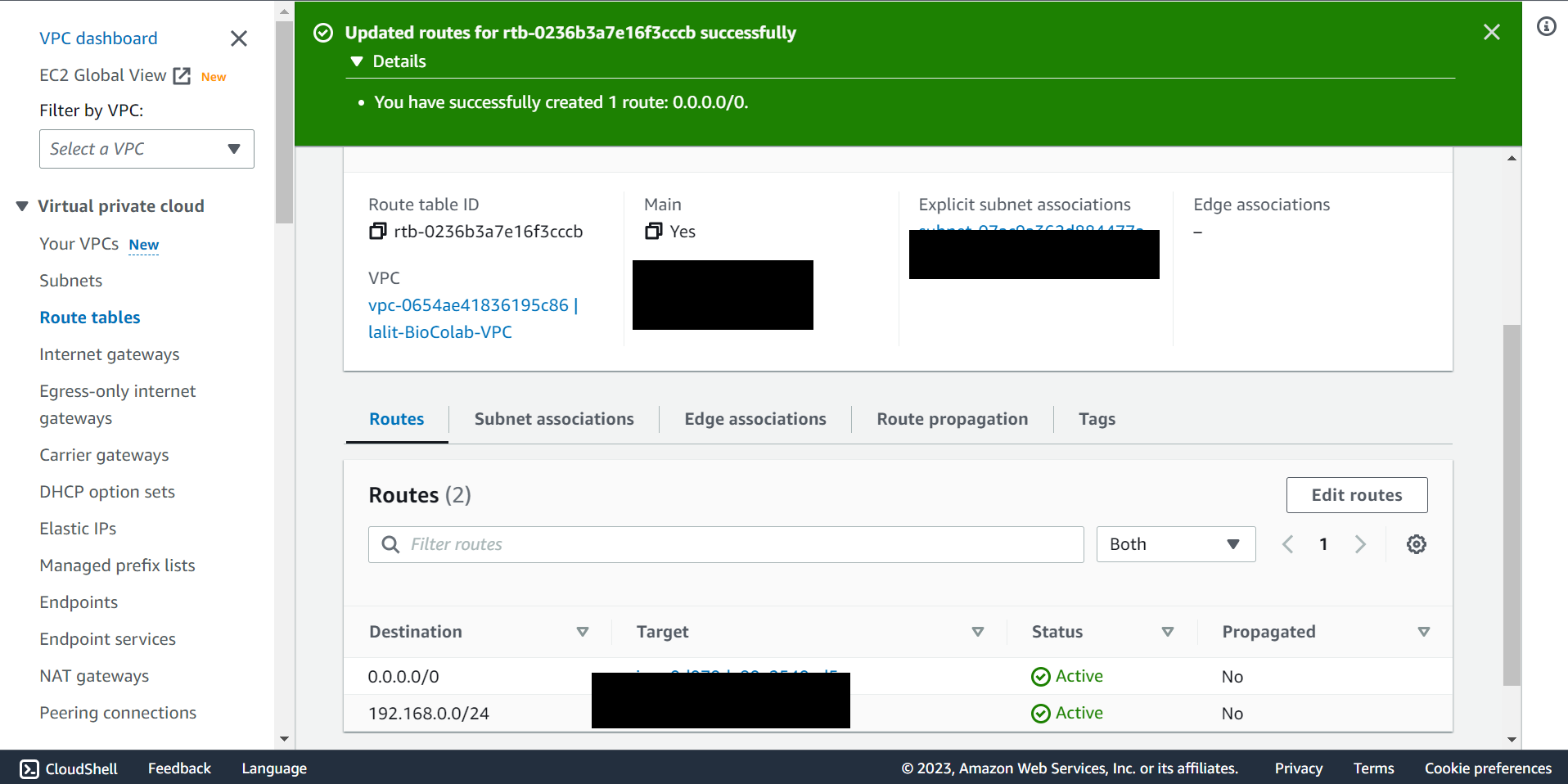
🔶 Router table updated.
Create ec2 Instance ¶
🔶 Click on Search Bar and type ec2.
🔶 Click on instances (running).
🔶 Click on Launch instances.
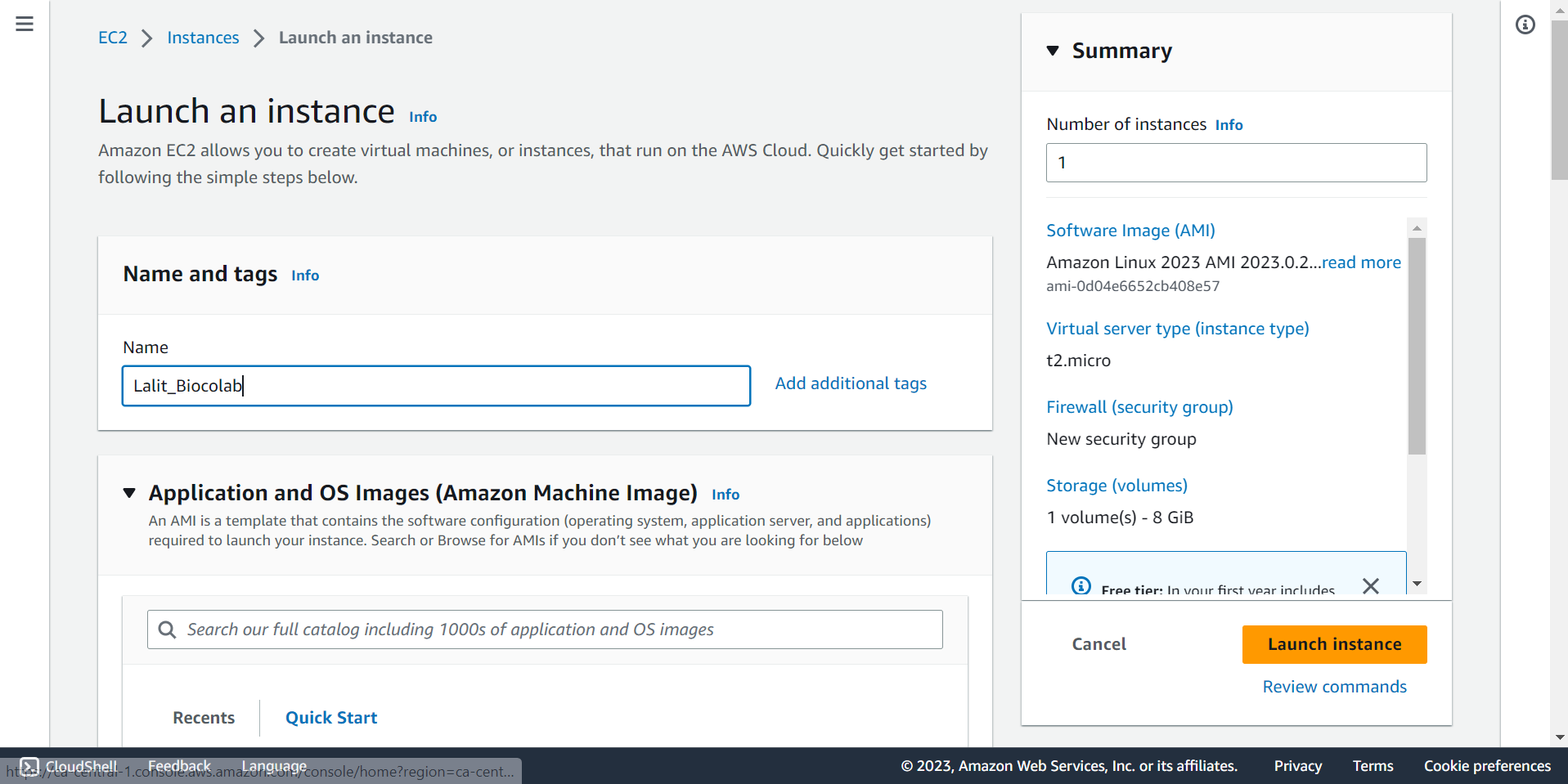
🔶 Type name of instance.
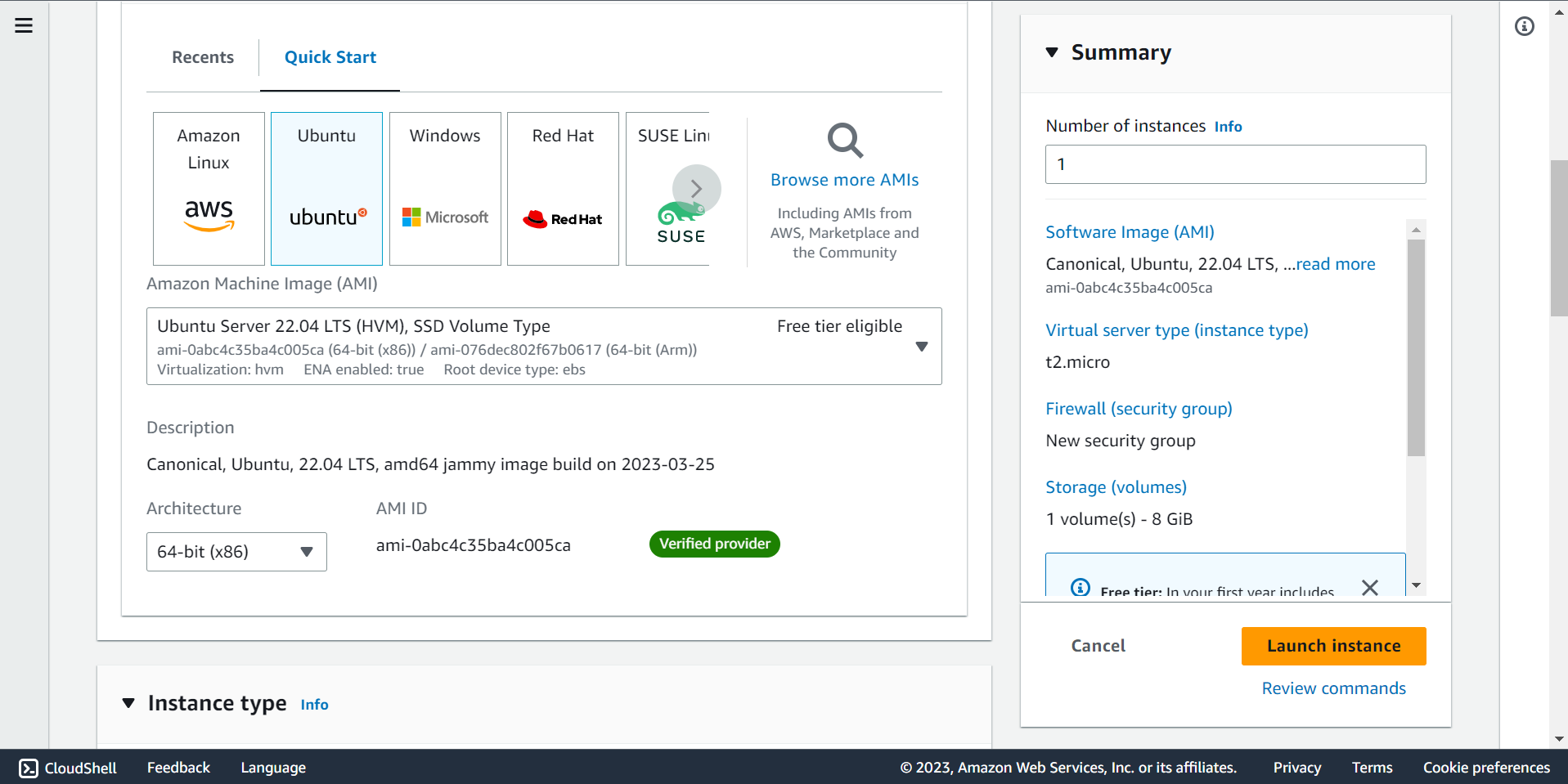
🔶 Select Operating System. Here, We are selecting ubuntu.
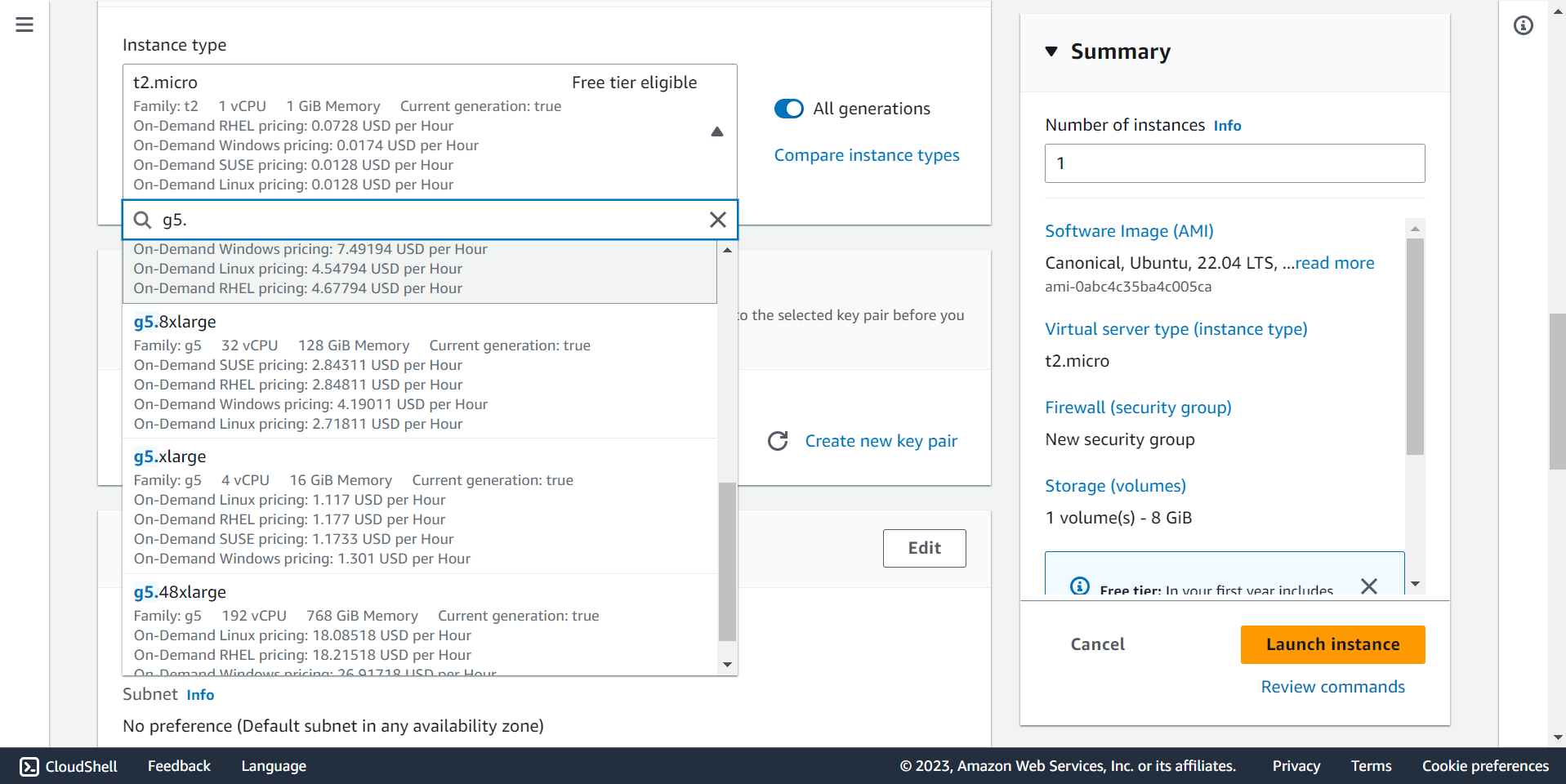
🔶 Select g5.8xlarge instance.
🔍 Select instance type based on your requirements.
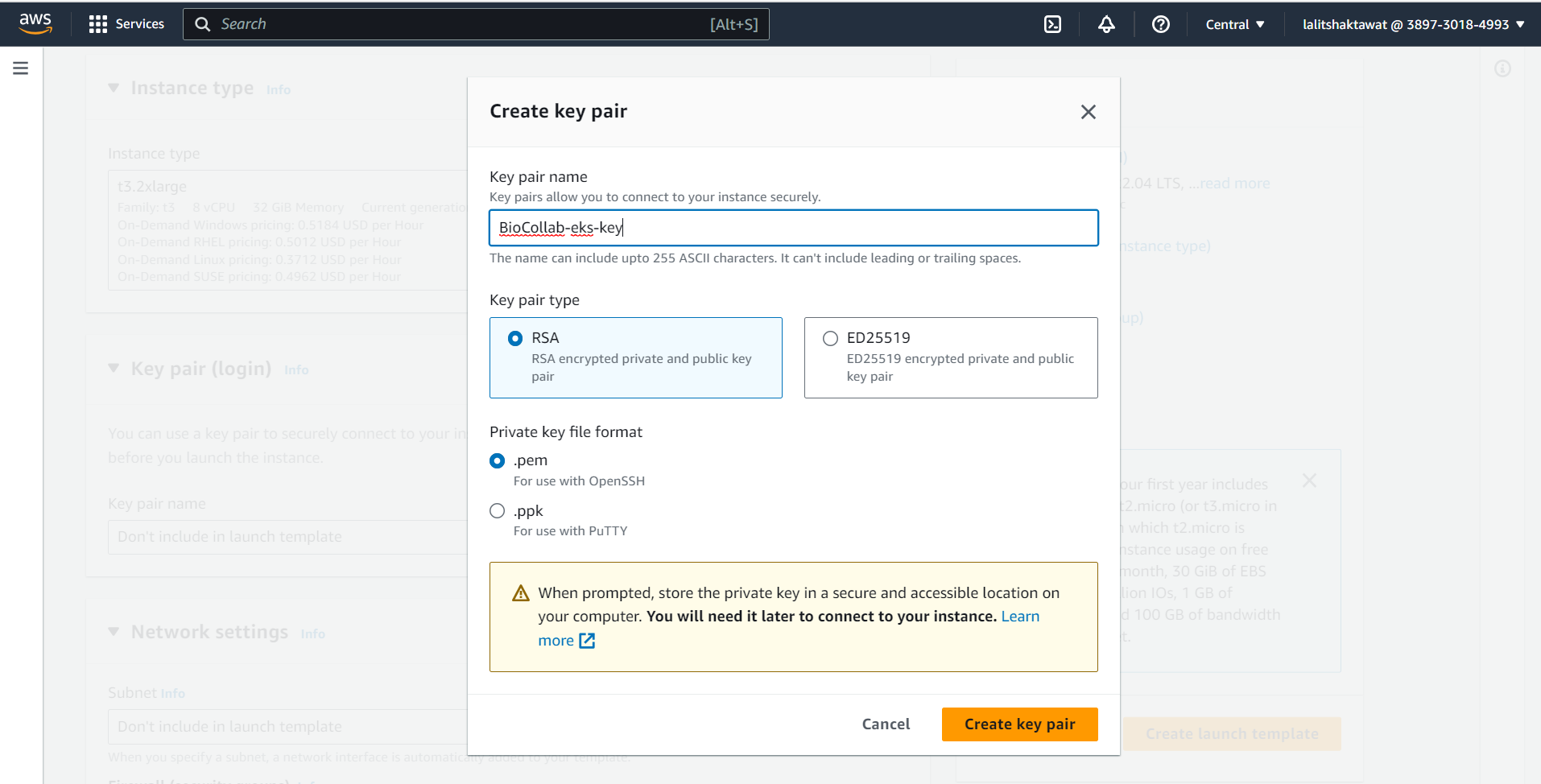
🔶 Create a Keypair to connect to ec2 instance. Type Key pair name and Click on Create key pair.
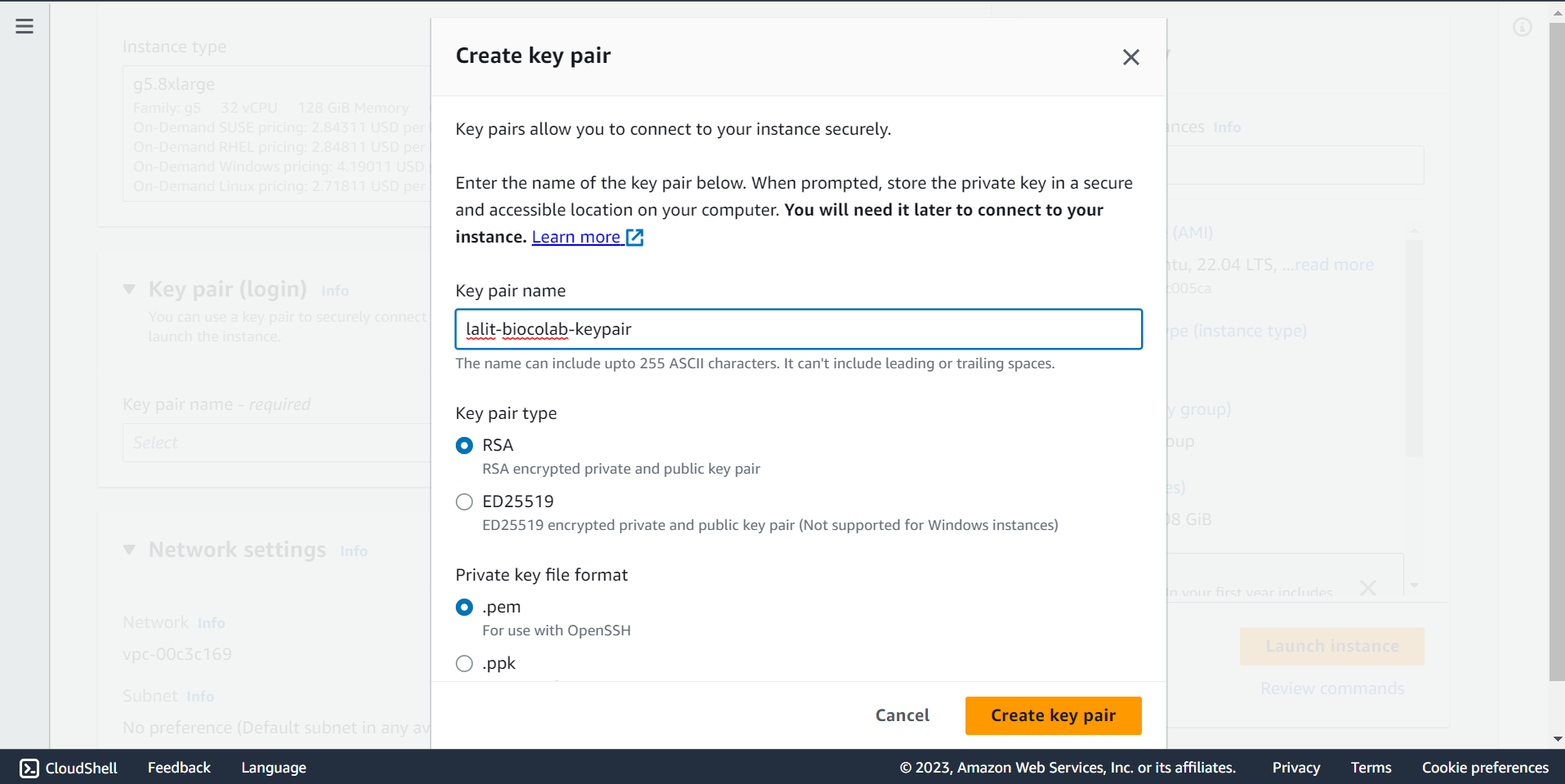
🔶 Key pair will download.
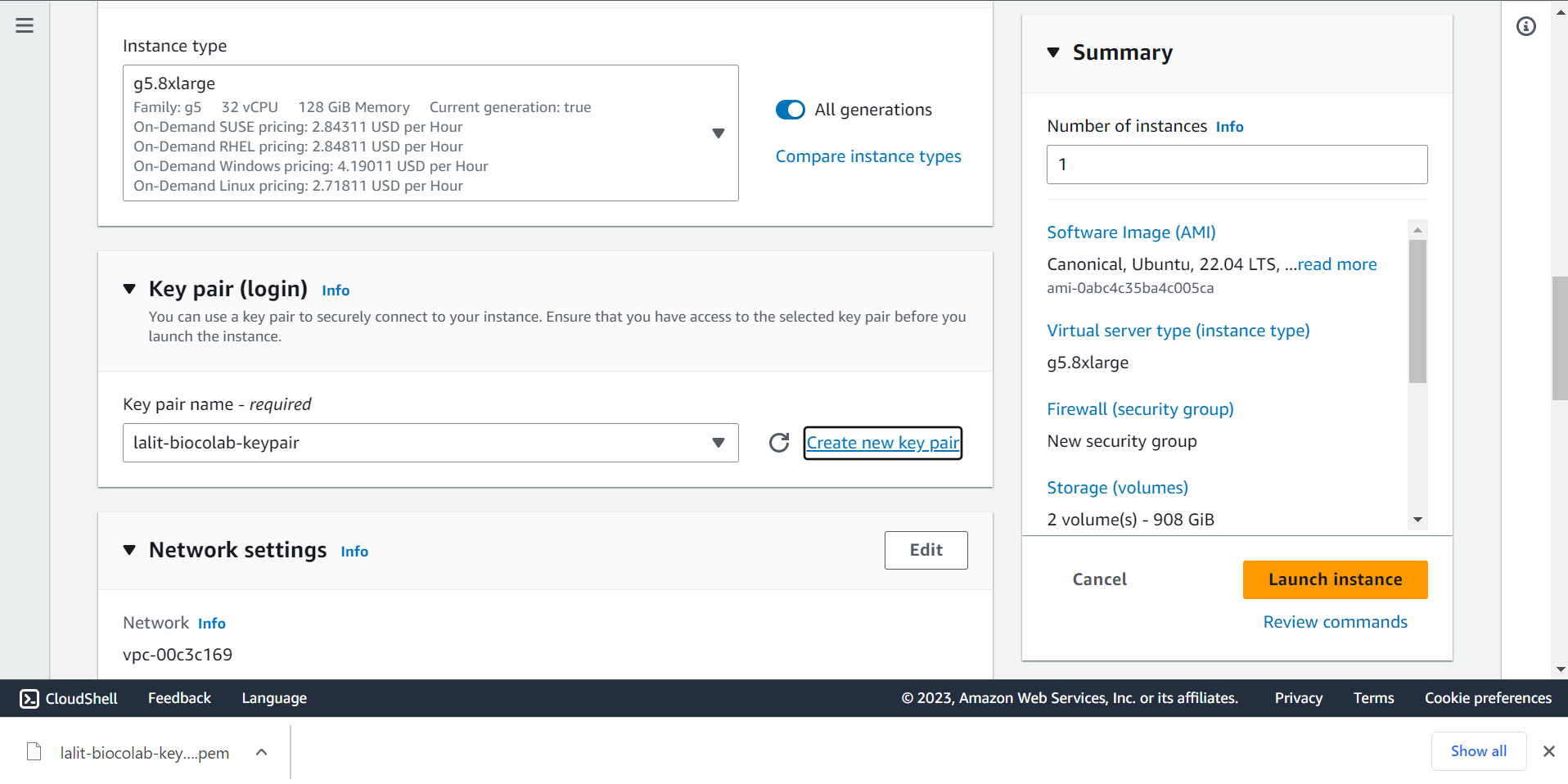
🔶 On the Network settings select correct VPC and Subnet ID
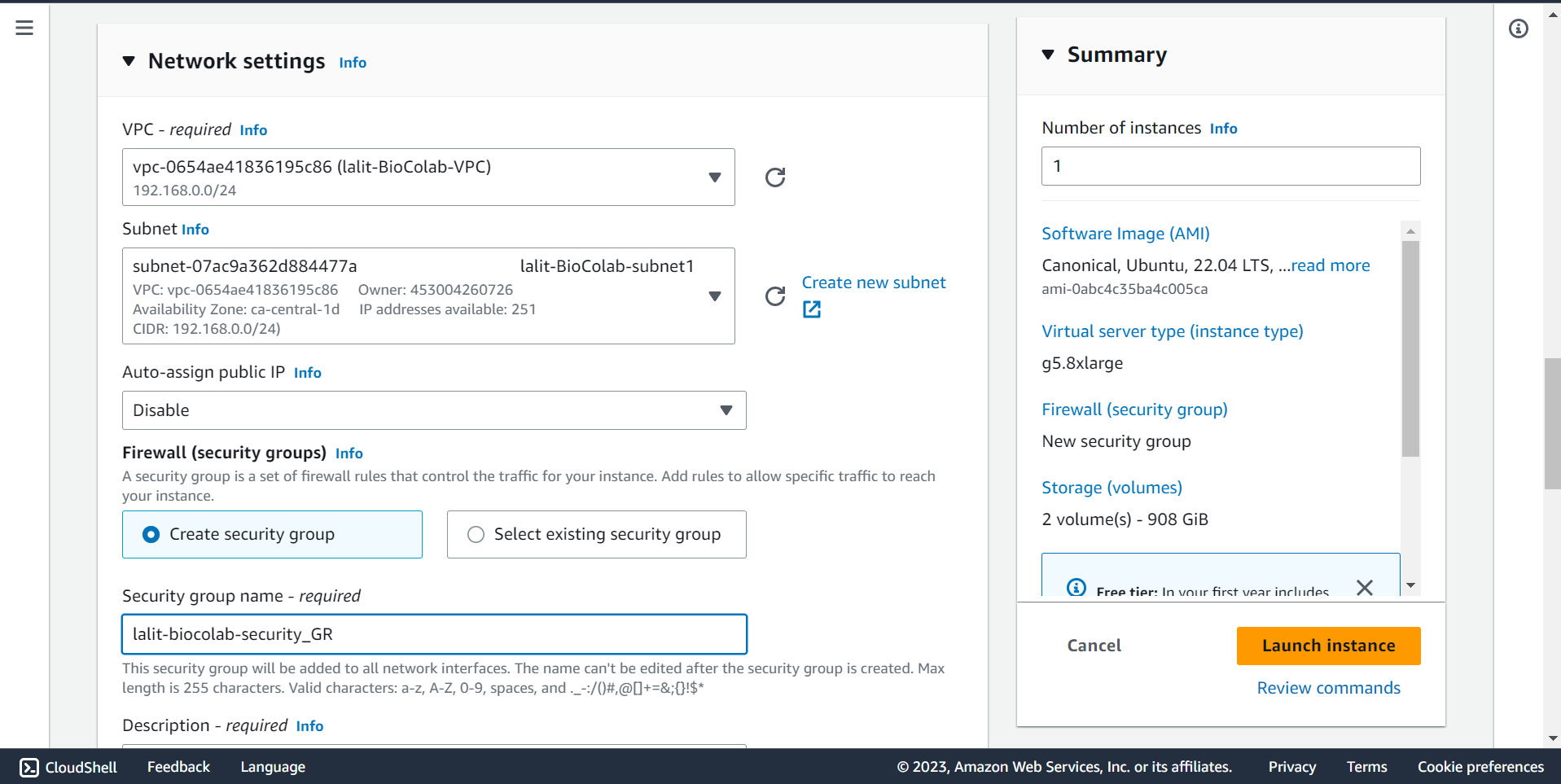
🔶 On the firewall (Security groups) section. Create new security group by giving appropriate name.
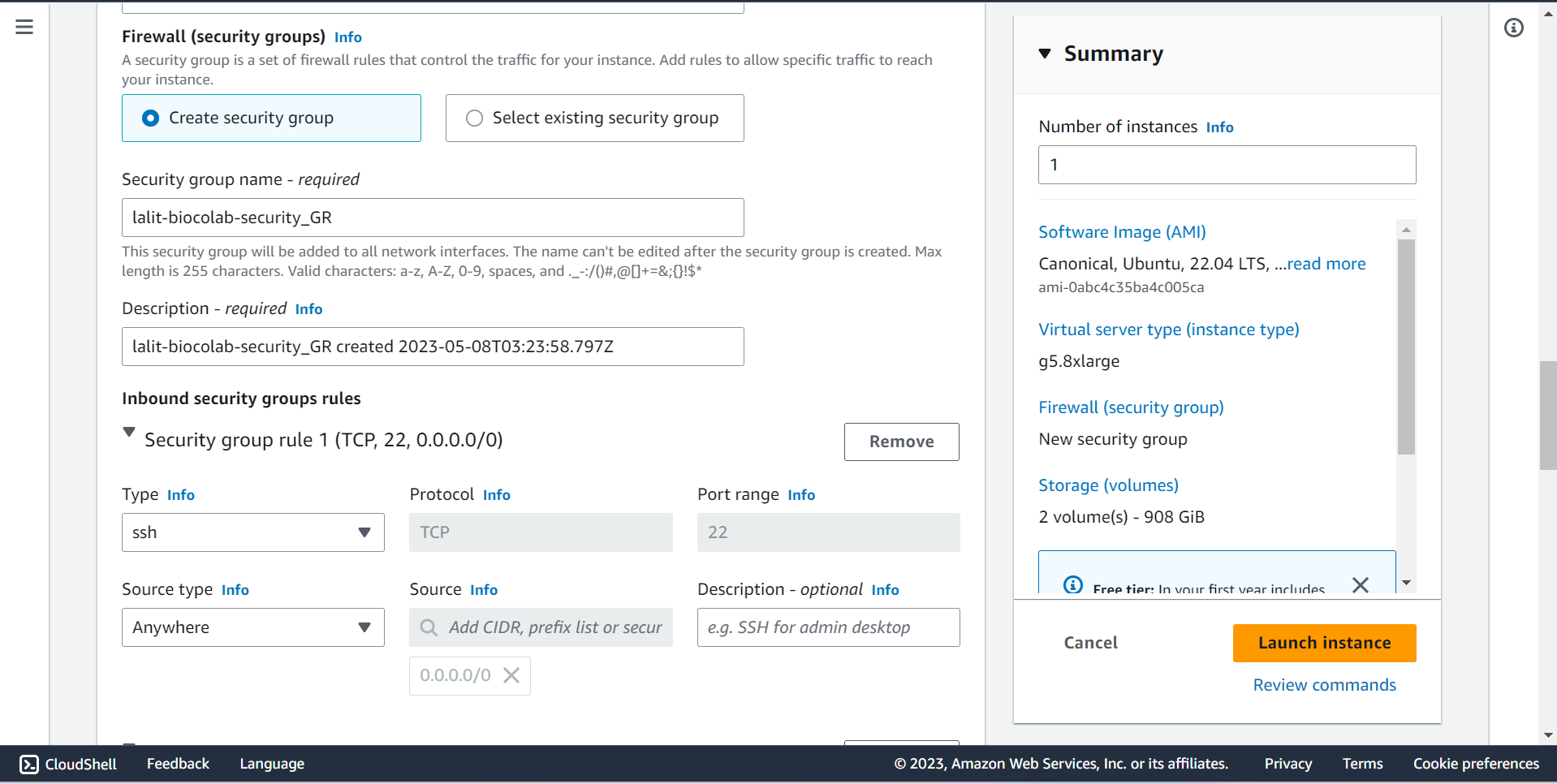
🔶 Allow SSH , HTTP and HTTPS. Source will be 0.0.0.0/0.
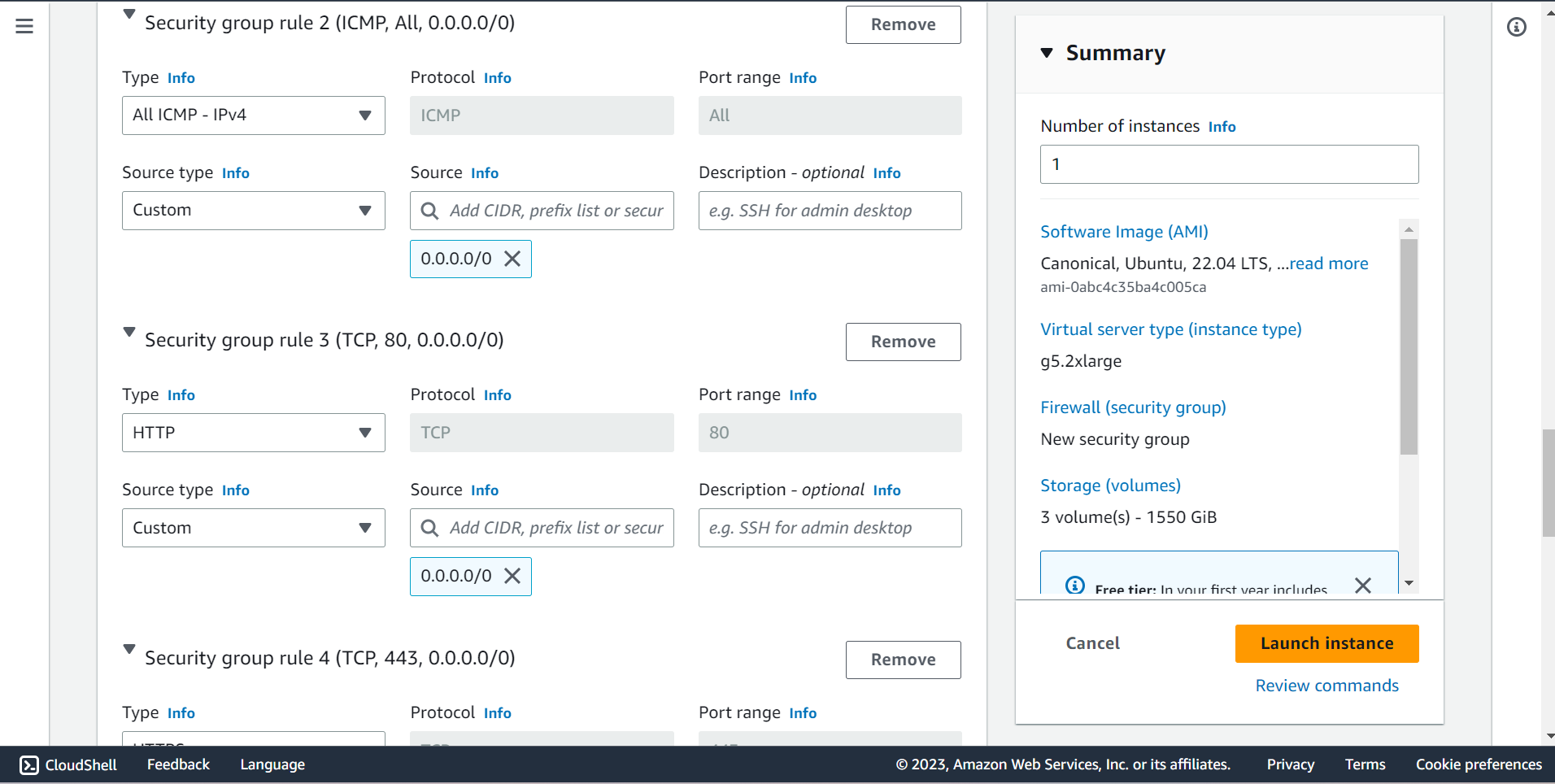
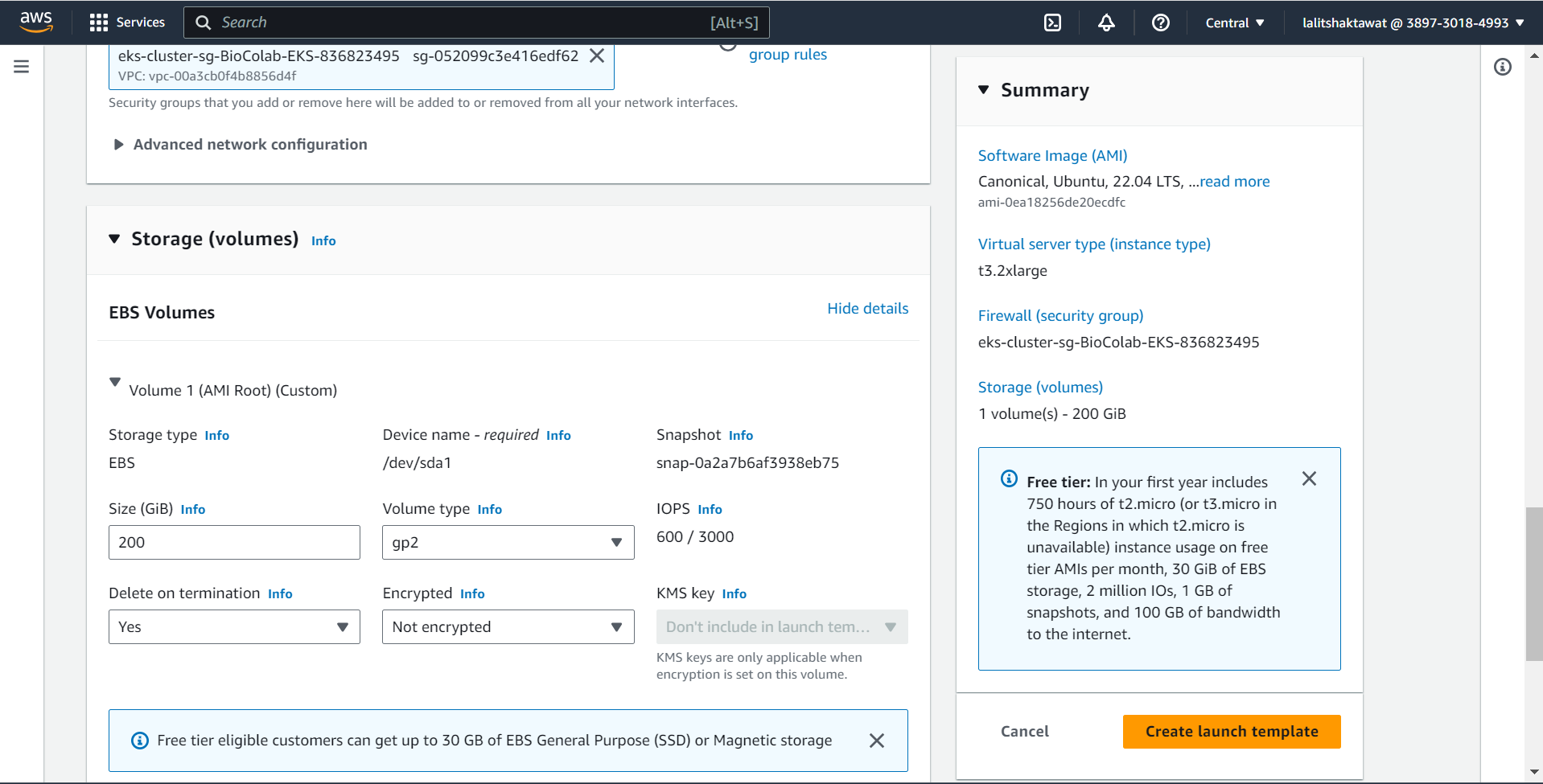
⭕ Configure Storage
🔶 Root Volume is 100GB
🔶 EBS Volume is 1000GB ~ 1TB
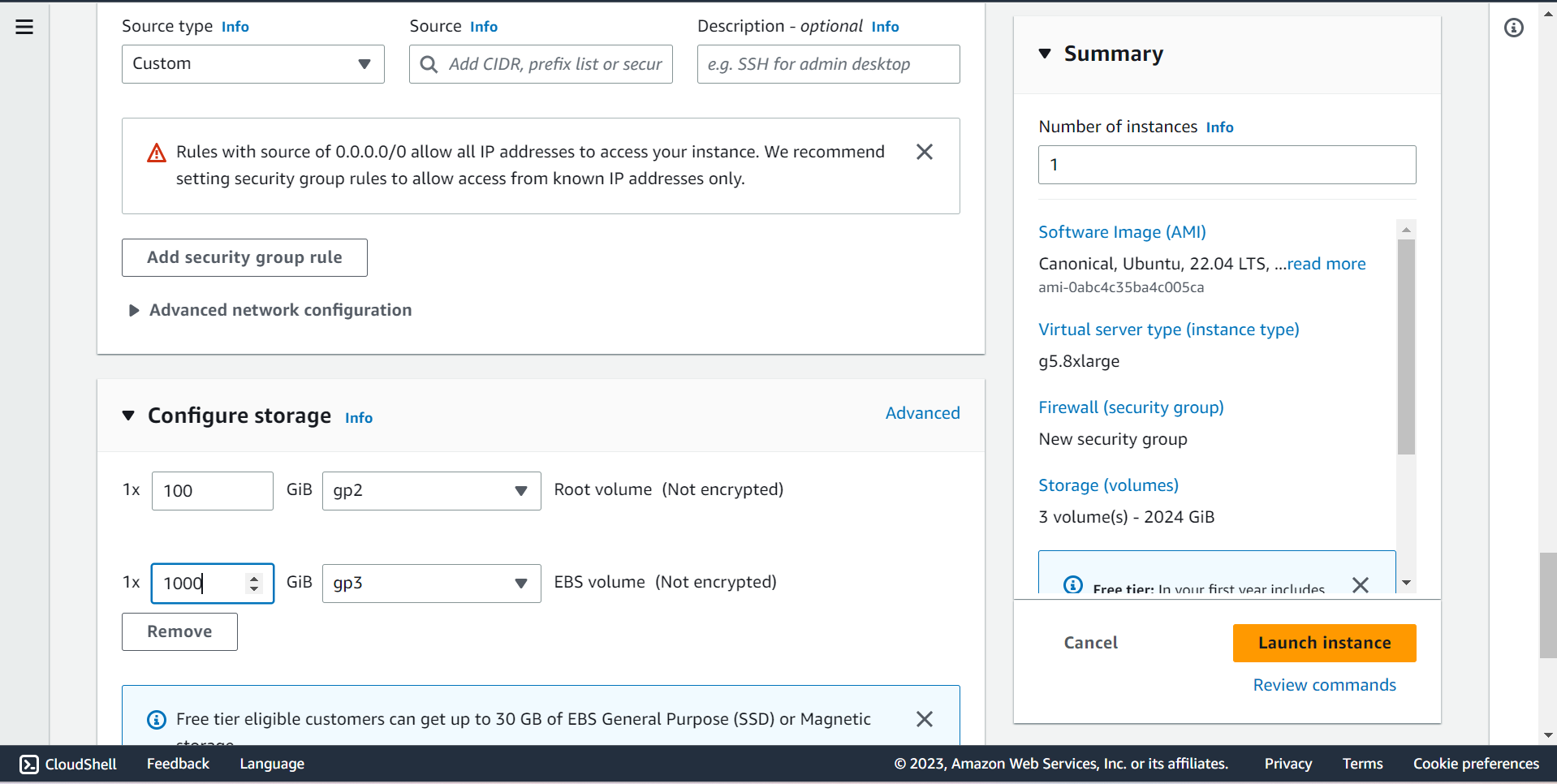
🔶 Click on Launch instance.
🔶 Instance creation will be in progress.
Assign Elastic IP to instance ¶
🔶 Click on Elastic IPs and Click on Allocate Elastic IP address.
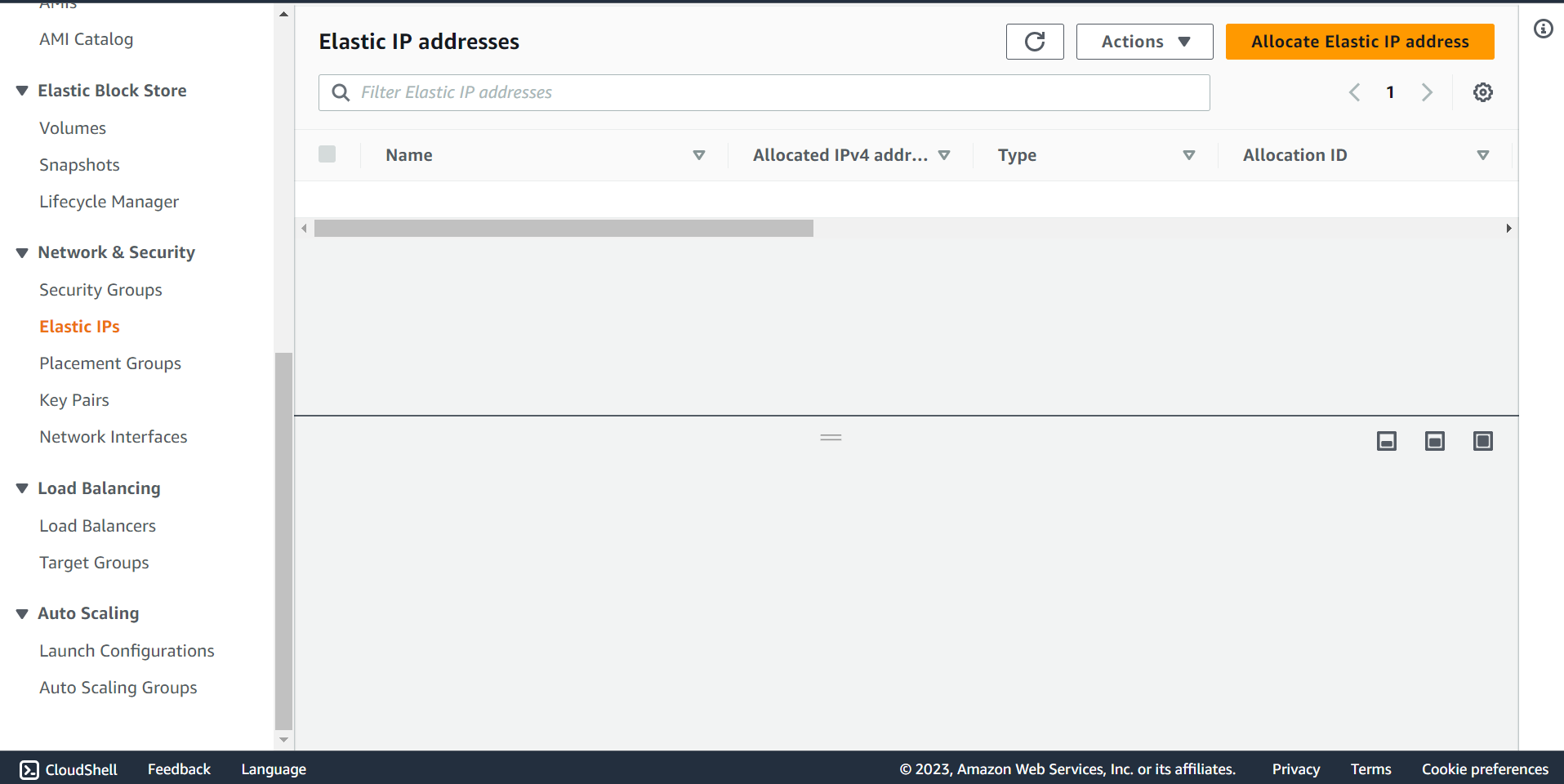
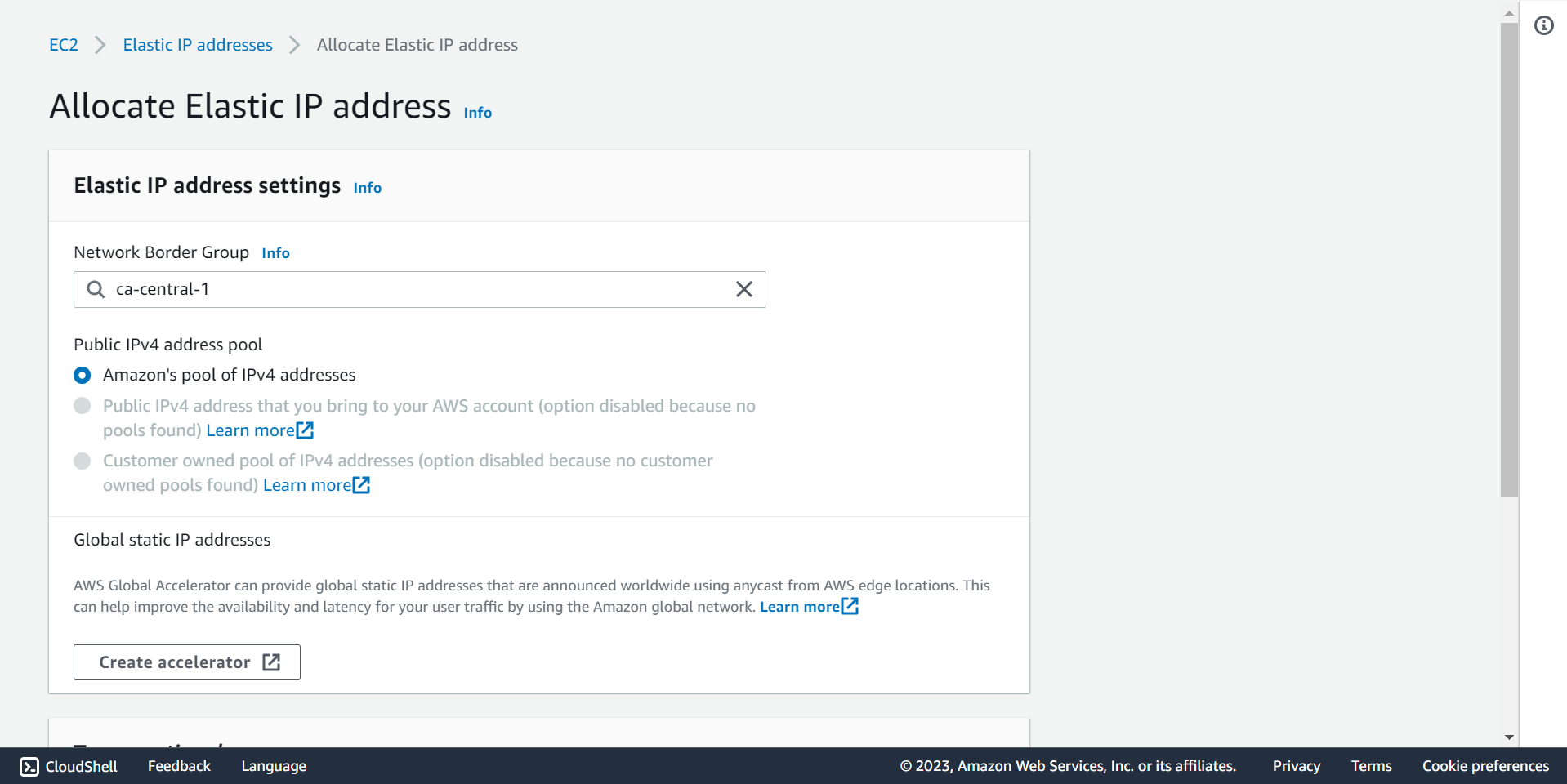
🔶 Select Network Boarder Group and take Elastic IP address.
🔶 Click on Associate Elastic IP address.
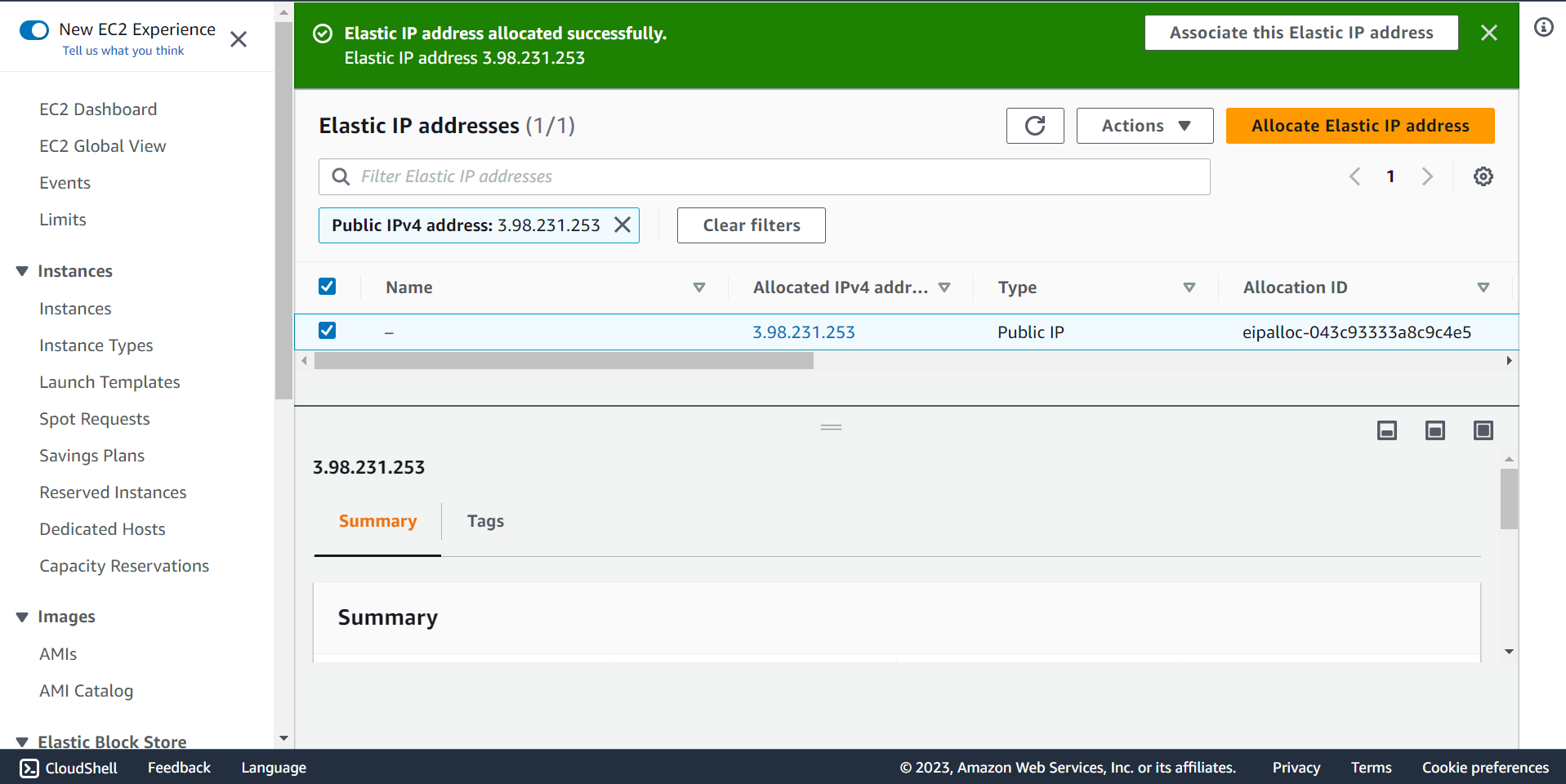
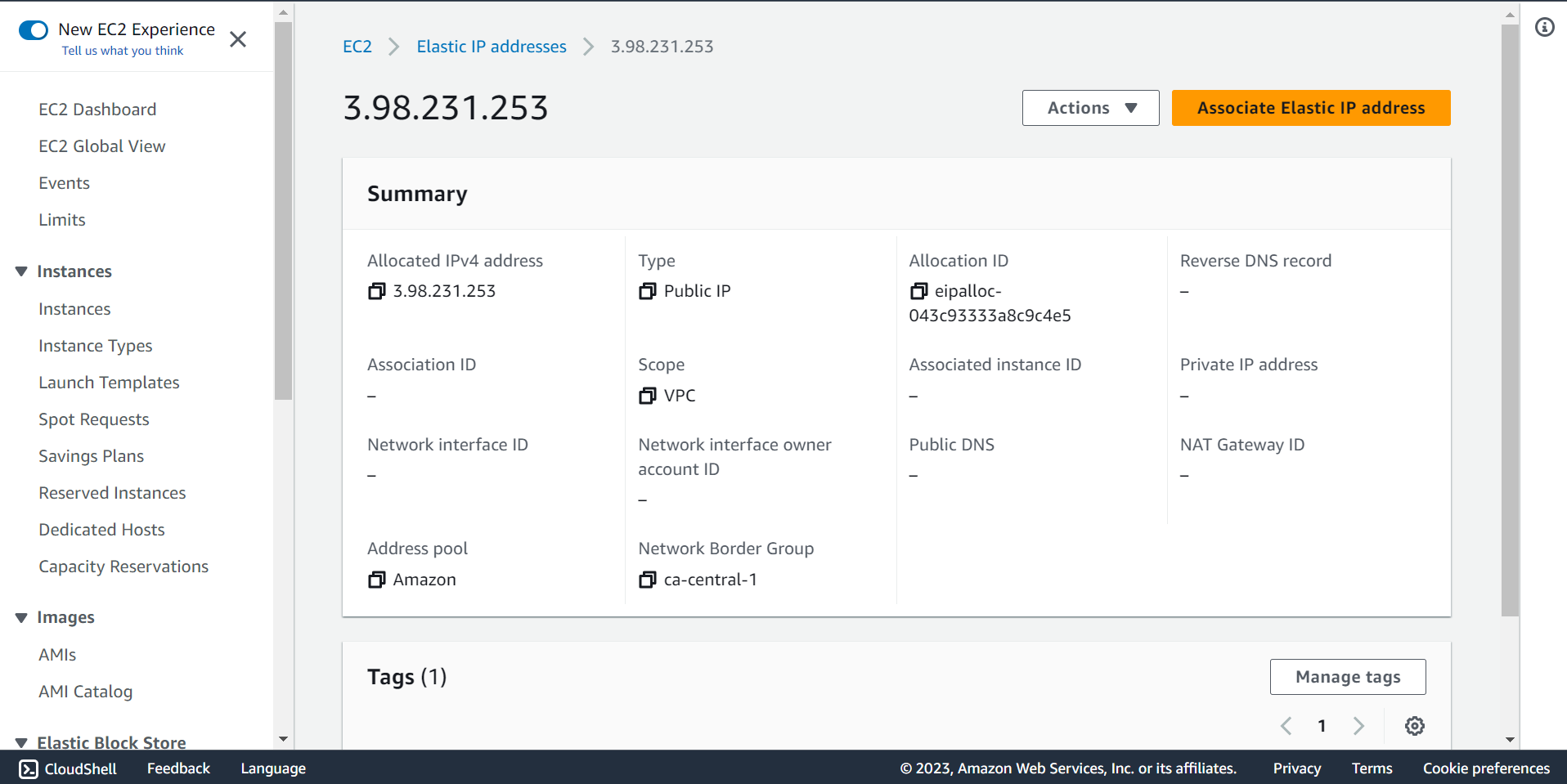
🔶 Select Instance and Private IP address.
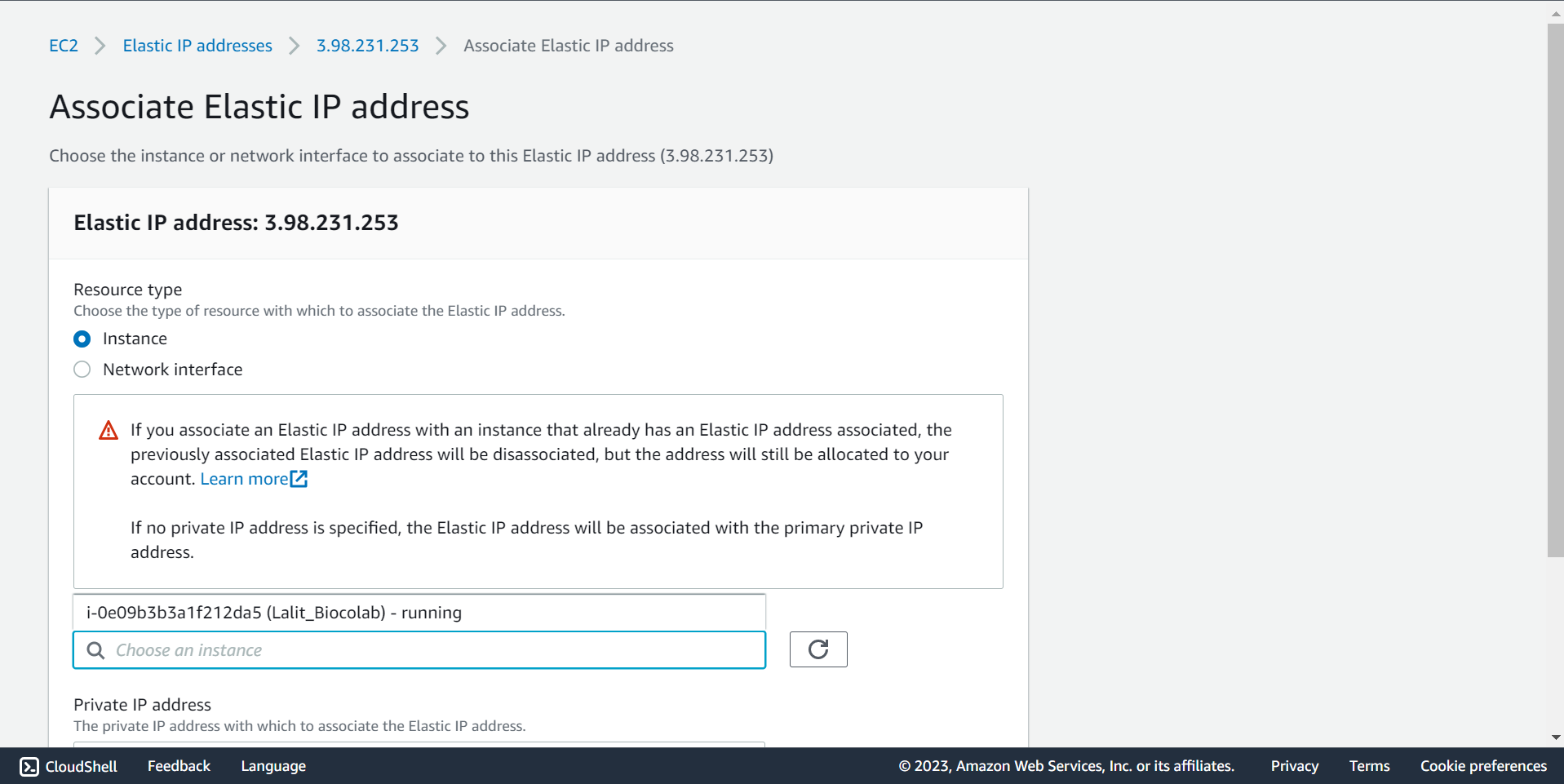
🔶 Click on Associate.
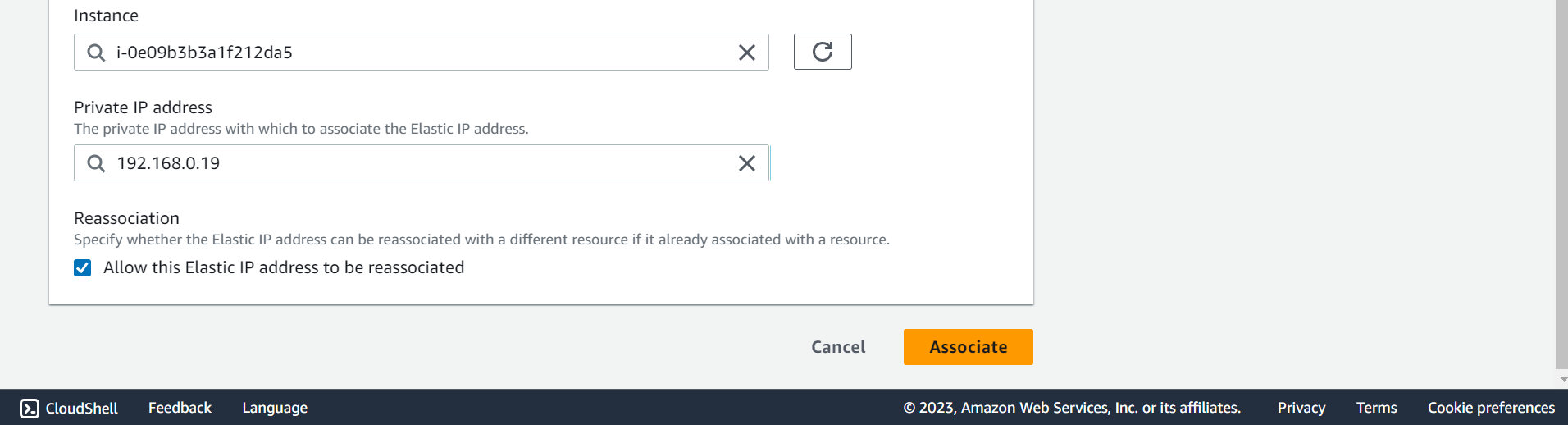
🔶 Elastic IP address associated successfully.
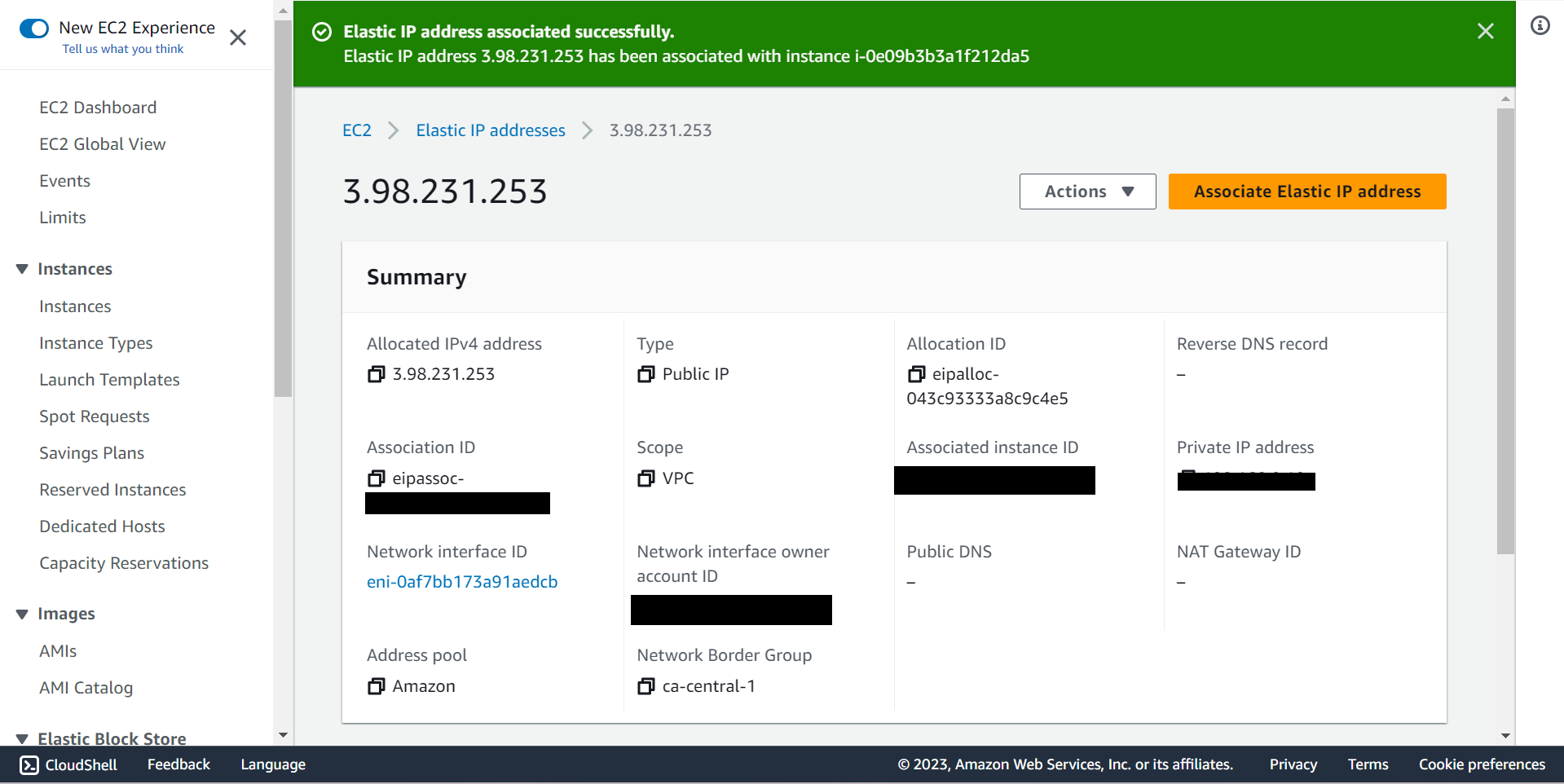
🔶 Take connection string and SSH to ec2 Instnace.
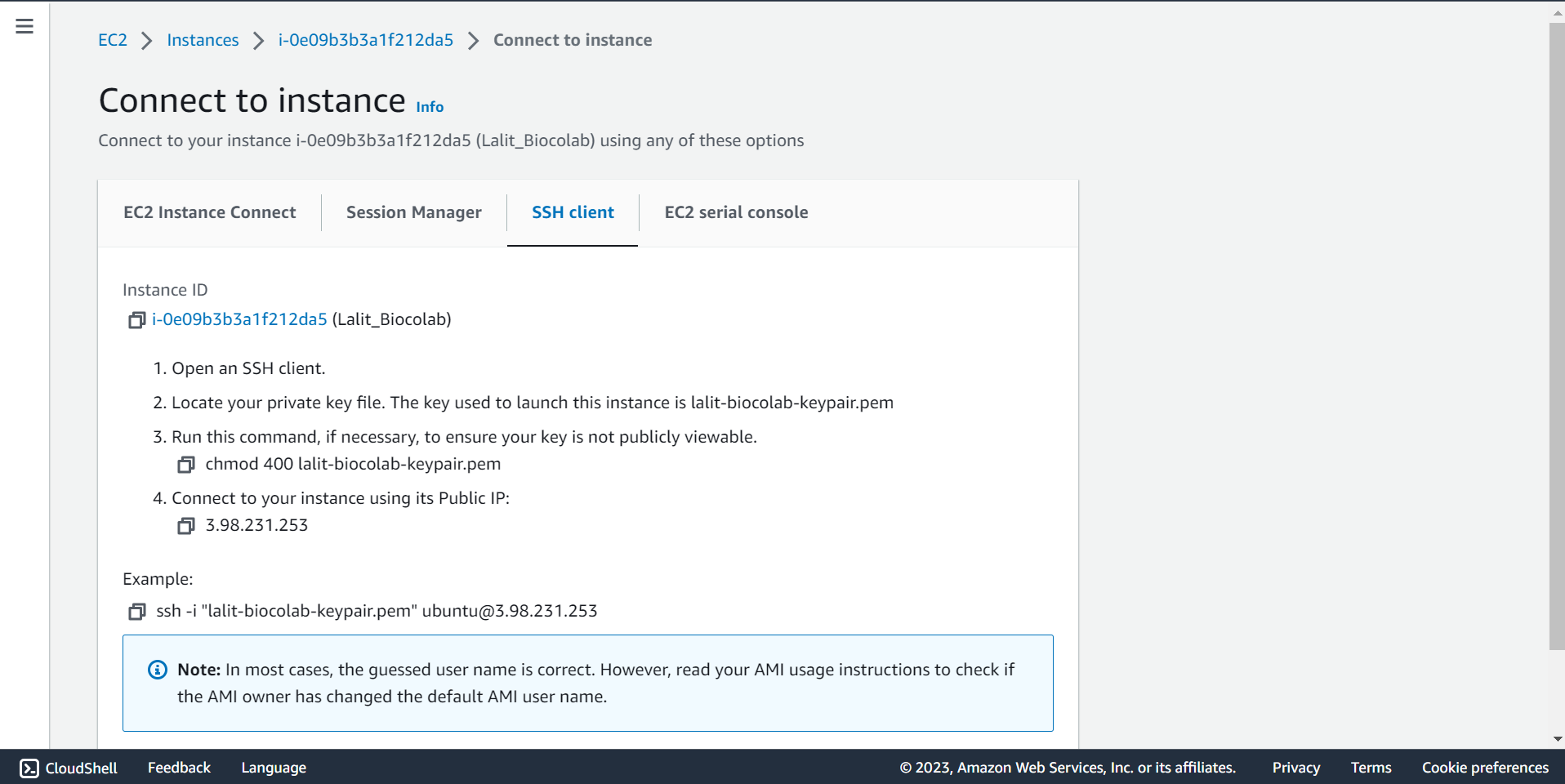
Installation Started. ¶
# Copy the connection string. Use SSH key pair and connect to ec2 instance.
ssh -i "lalit-biocolab-keypair.pem" ubuntu@3.98.231.253
# Before start installation. Kindly create below path.
| Item | Note | Size |
|---|---|---|
| BioStudio Token | To access our product. | |
| Application Domain | Access BioStudio on Browser. | |
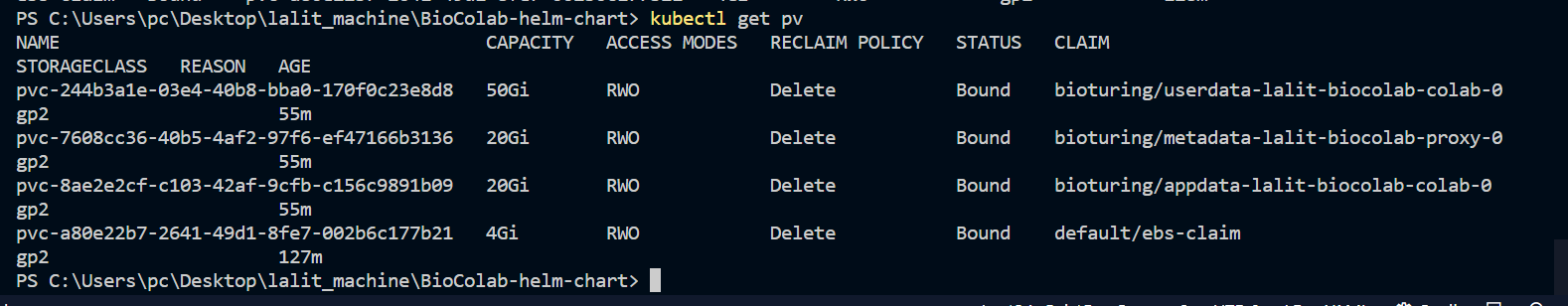
| META_DATA Volume | This will use by Bioproxy to store database. [/biocolab/metadata] | 50GB |
| SSL Volume | Using by BioProxy. [/biocolab/configs] | 1GB or above |
| Data Volume | Using to store user data. [/biocolab/userdata] | 500GB or above |
| Application data Volume | Used to store application binary data.[/biocolab/appdata] | 150GB or above |
| Ethernet IP Address | Use to pass IP address during installation (eth0) |
Run the install.biocolab.docker.sh script for docker environment and install require software.
# Create required folder structure before execution.
mkdir -p /biocolab/metadata
mkdir -p /biocolab/configs
mkdir -p /biocolab/userdata
mkdir -p /biocolab/appdata
# Switch to /biocolab folder.
cd /biocolab
# Download script:
# Note: Installation script version would be changed based on updates.
# Our Team will get you updates and keep in touch with you during installation.
wget https://github.com/bioturing/installation/archive/refs/tags/v2.1.11.tar.gz
# uncompressed .gz
tar xvf v2.1.11.tar.gz
# Switch to installation folder
cd installation-2.1.11/
# Execute installation script
bash install.biocolab.docker.sh
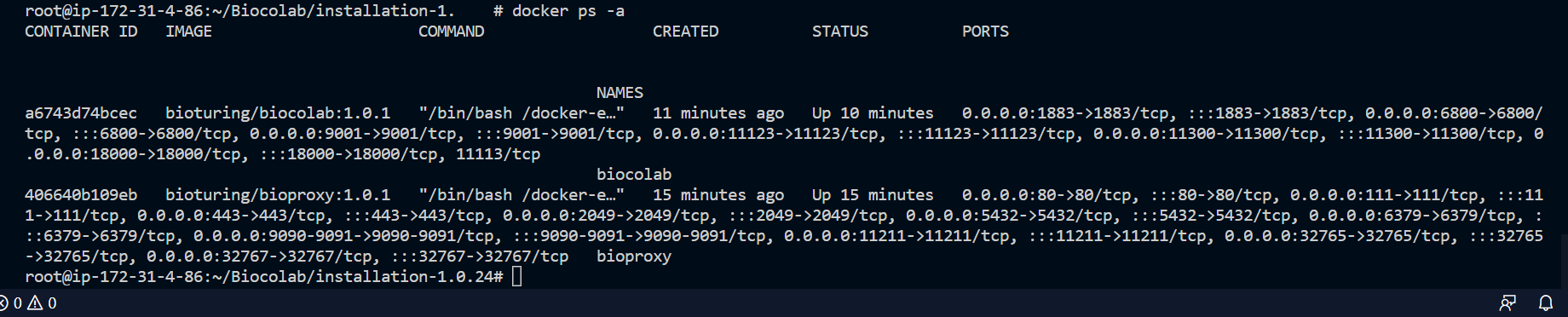
# Verify both container up and running.
docker ps -a
# Configure SSO login
http://<Your Domain>/dashboard/
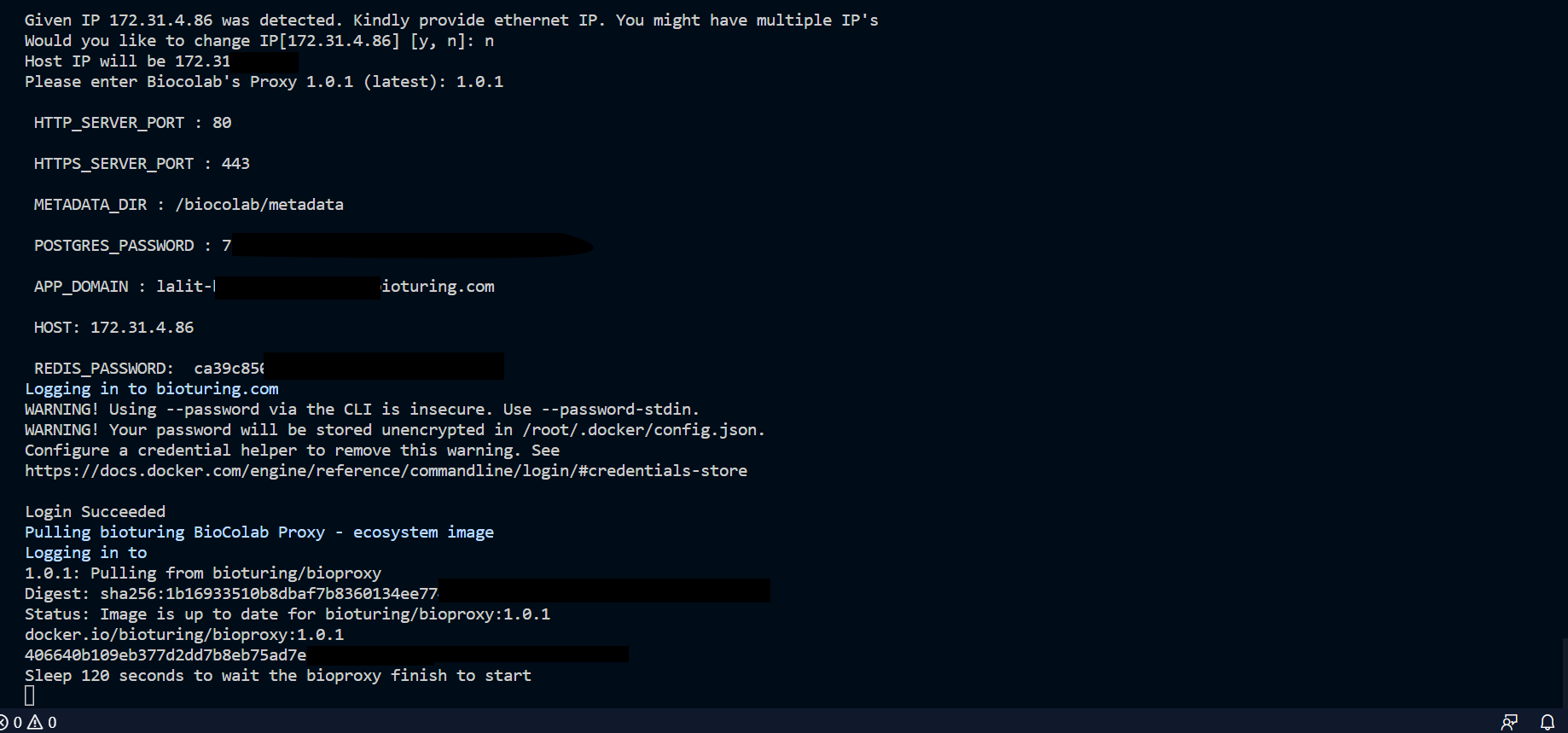
🔶 Download v2.1.11.tar.gz, which content script to install BioStudio.
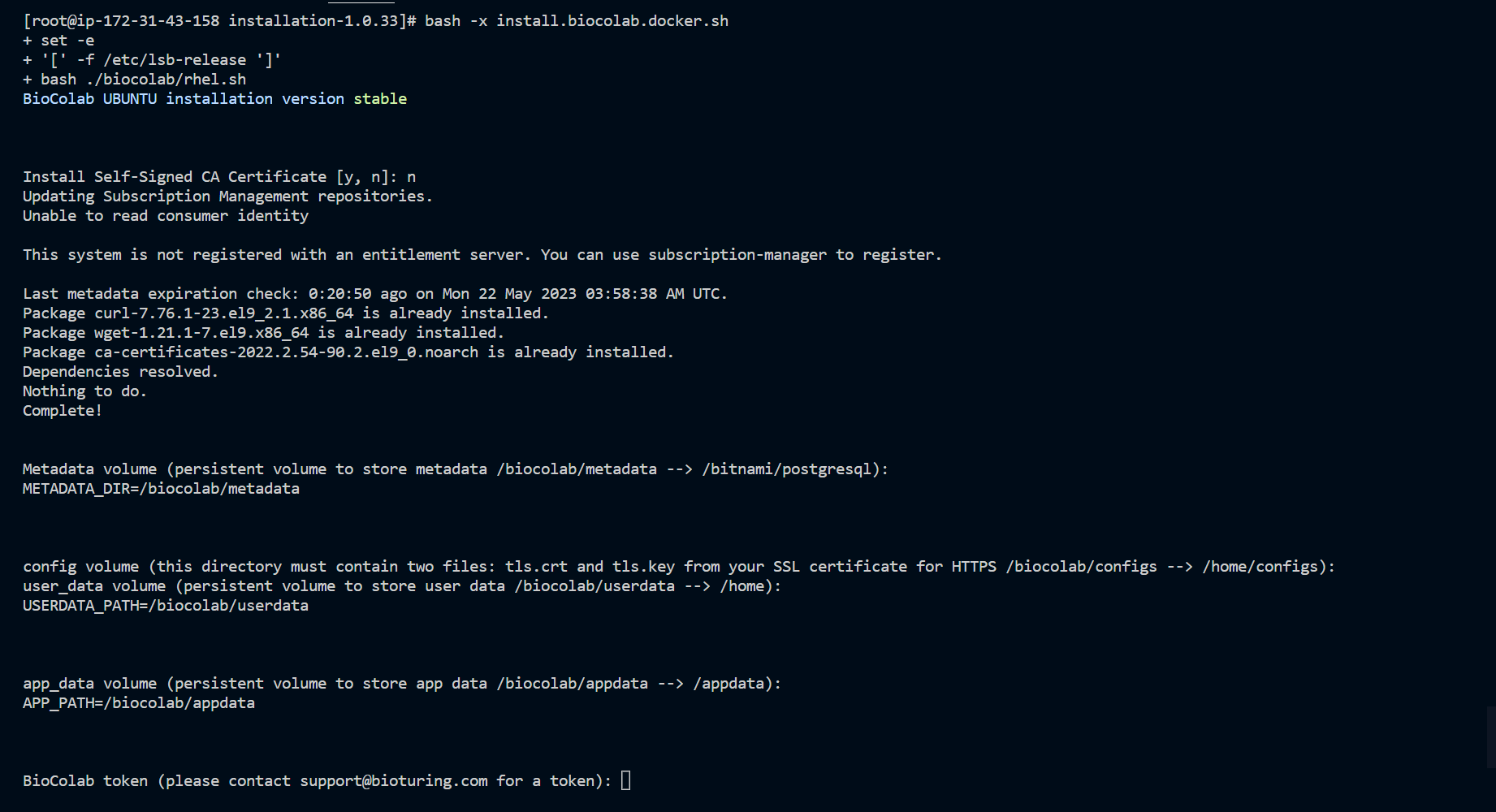
🔔 During execution of this script, It will prompt for input credential. Kindly note that user name is admin and keep store the password safely, which you are going to input. It needed to be performed admin activities.
🔶 Execute script.
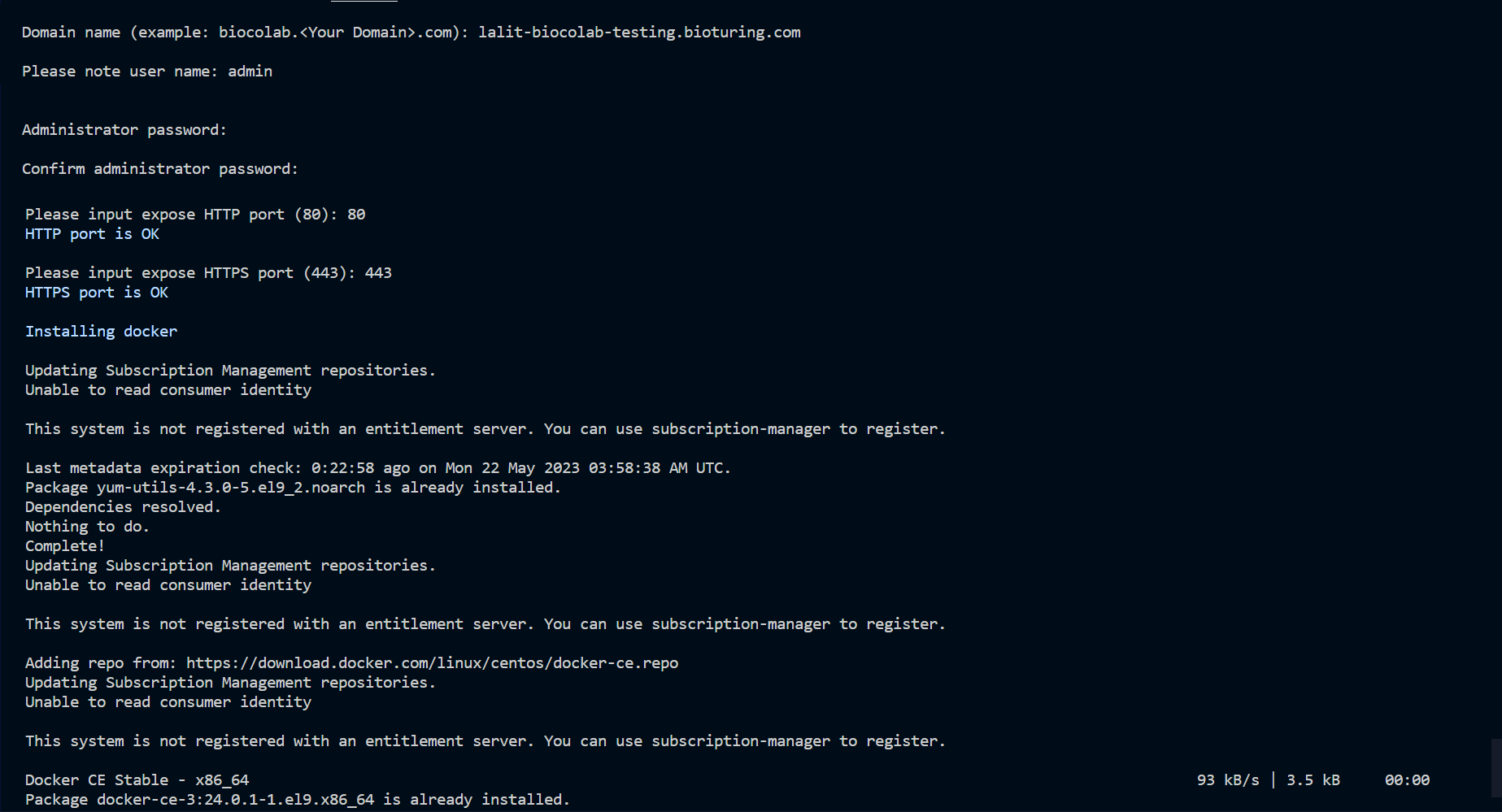
🔶 Follow execution script’s instructions and input values accordingly.
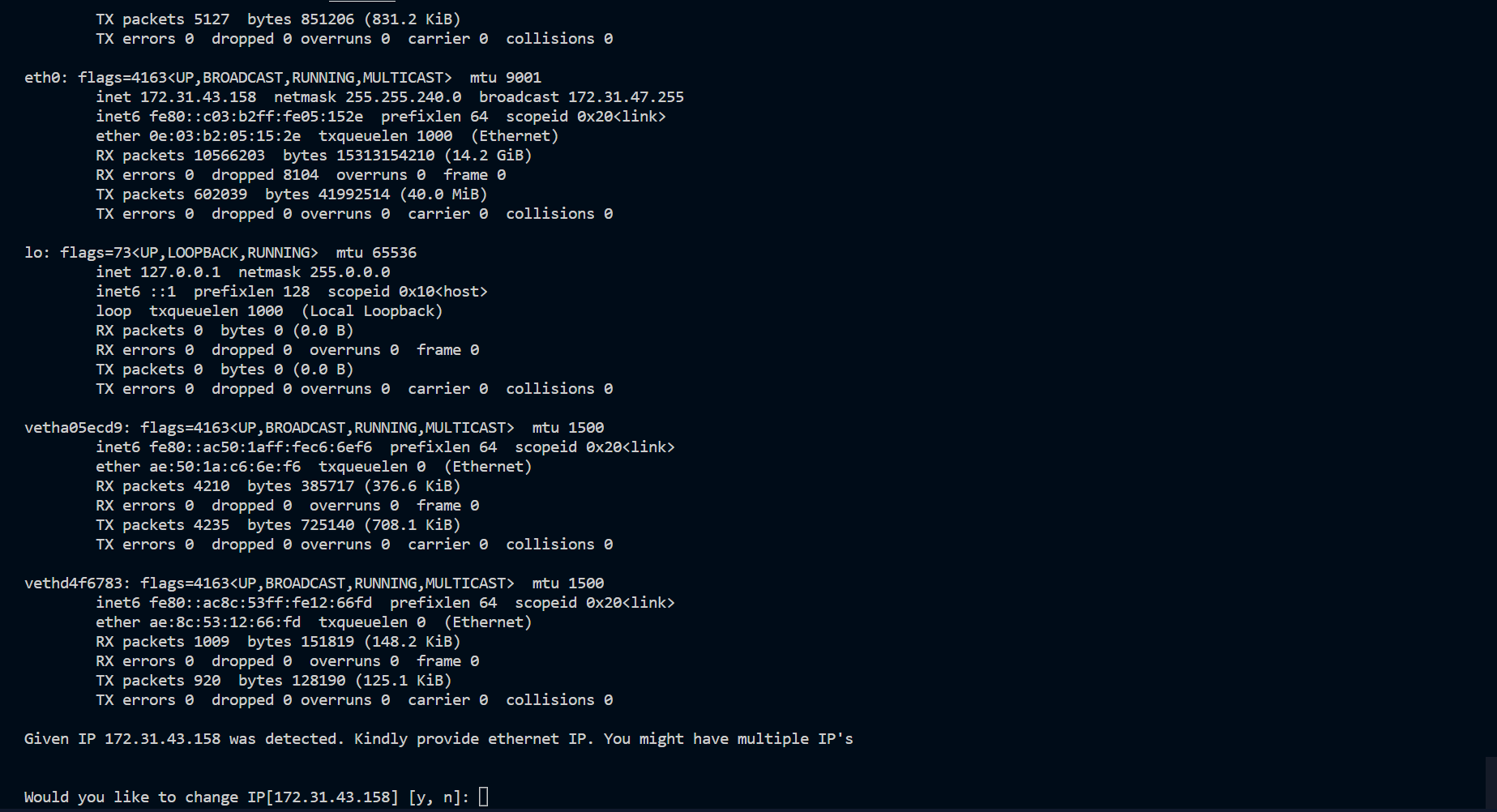
🔶 Change the IP, If we have many.
BioProxy version : 1.0.26
BioColab version : 2.0.50
# Please provide the version based on Bioturing instruction.
# It would be vary based on updates.
🔶 Verify both container status - UP and running.
🔶 Browser BioStudio.
# https://<Domain name>
⌚ Kindly wait for a while. BioStudio software will take time to download packages and install.
Register for root admin. ¶
https://<your domain>/dashboard
# It will prompt for Admin Login.
# Select Account Registration.
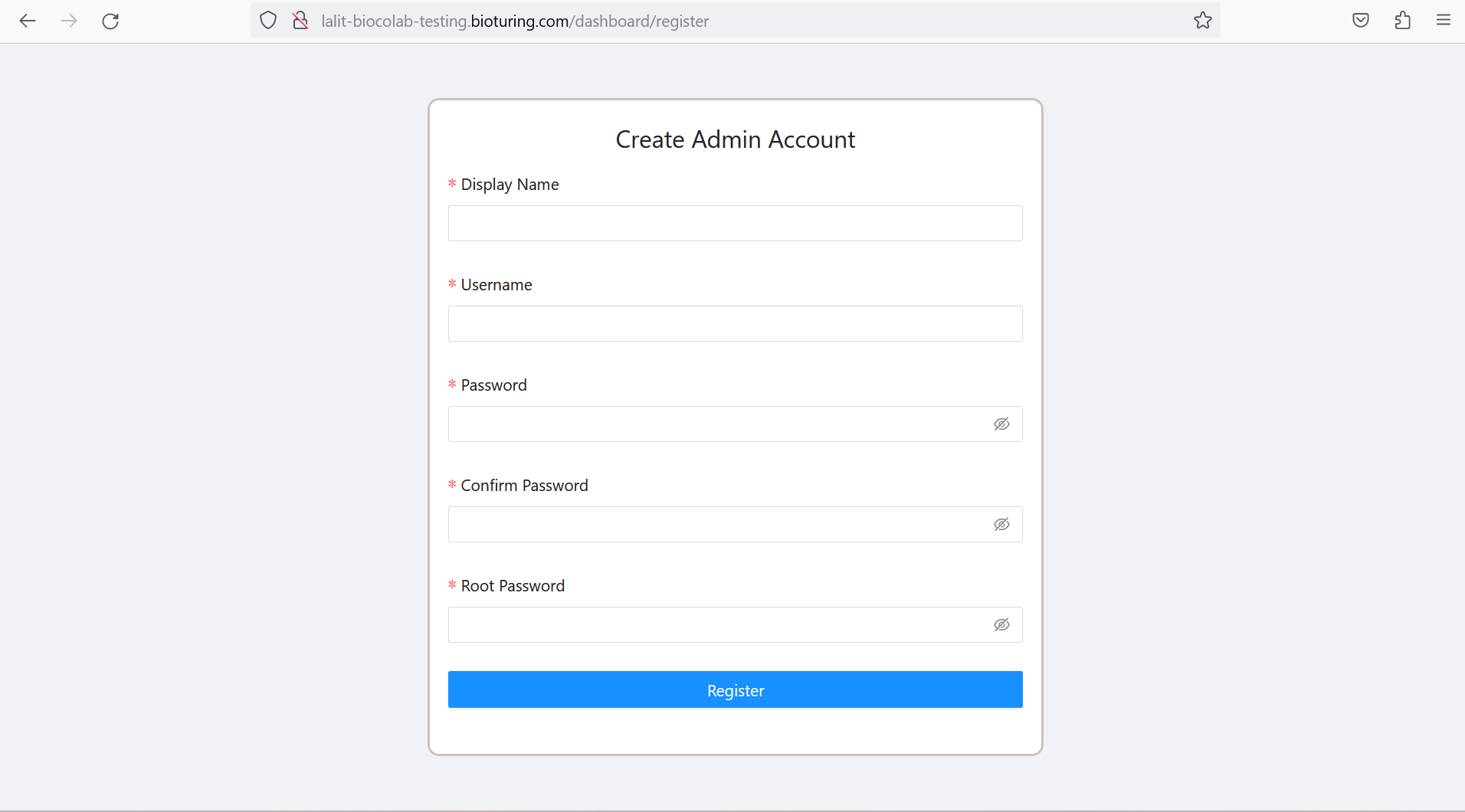
# Create admin account.
# Root Password is the one, we provided during installation.
# Login with the Dashboard using that account you registered earlier.
🔶 Account Registration.
🔶 Crearte Admin Account.
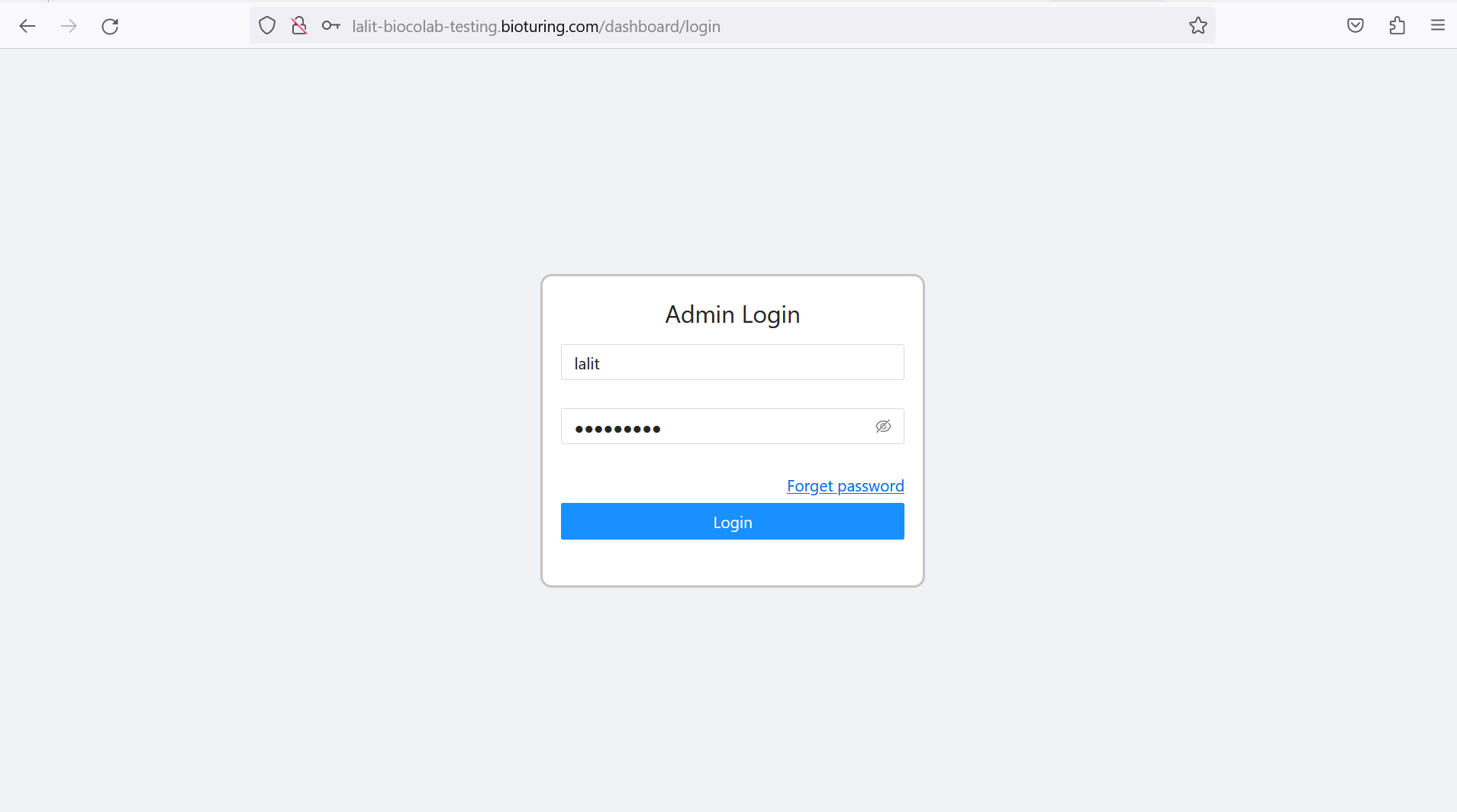
🔶 Login to dashboard.
https://<your domain>/dashboard
Workspace It is essential to create a machine and mount the volume before using the workspace.
Machine creation ¶
# Login with Dashboard using admin credential.
# Select Machines Tab form left-side menu.
# Select + Add New Machine.
Name: You can select any name to this machine. This is a private IP of your machine, That can be find in /etc/hosts file on BioStudio pods.
I already added a machine, so I will go with the update machine.
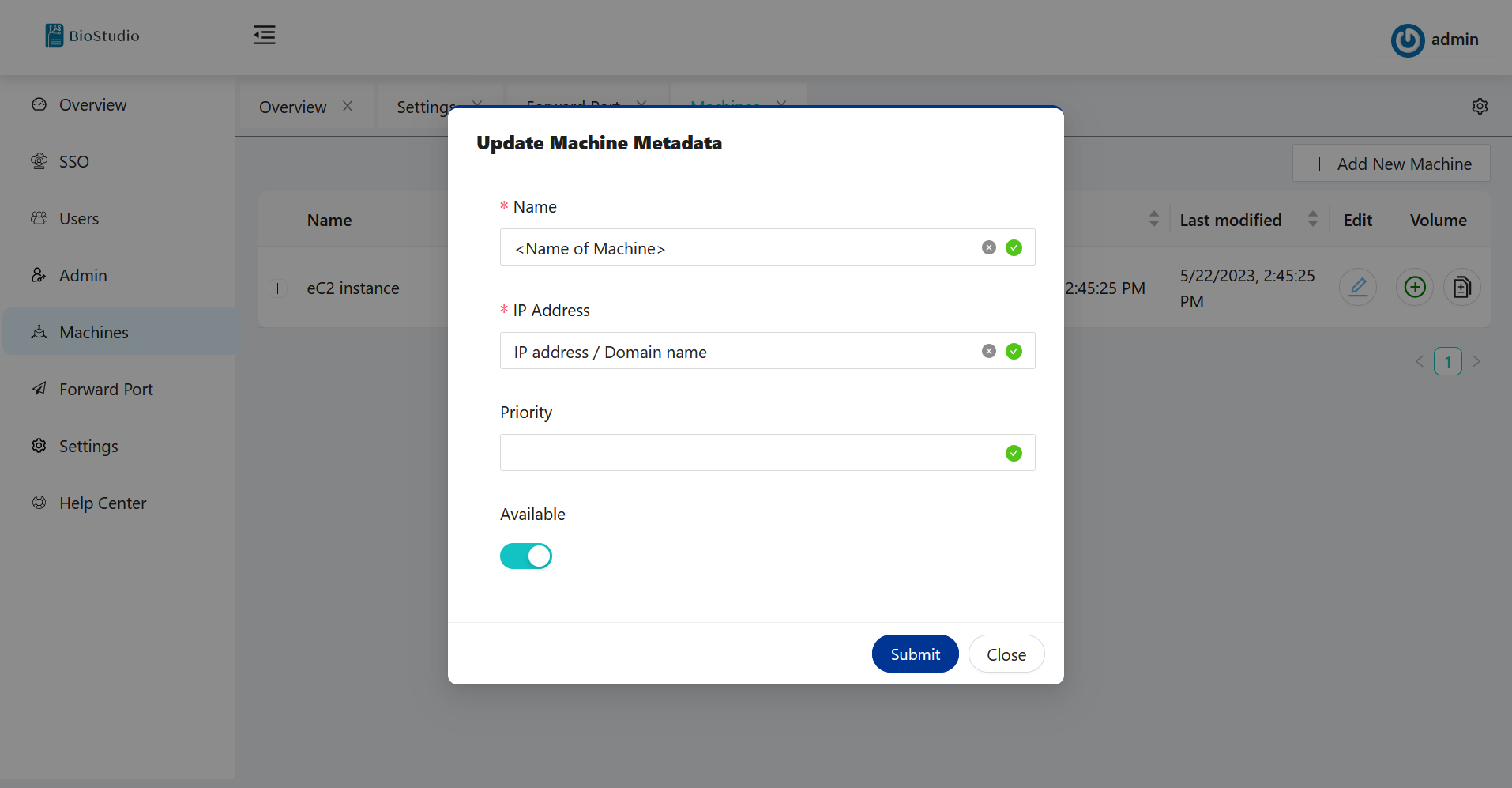
🔶 Please fill out Update machine metadata.
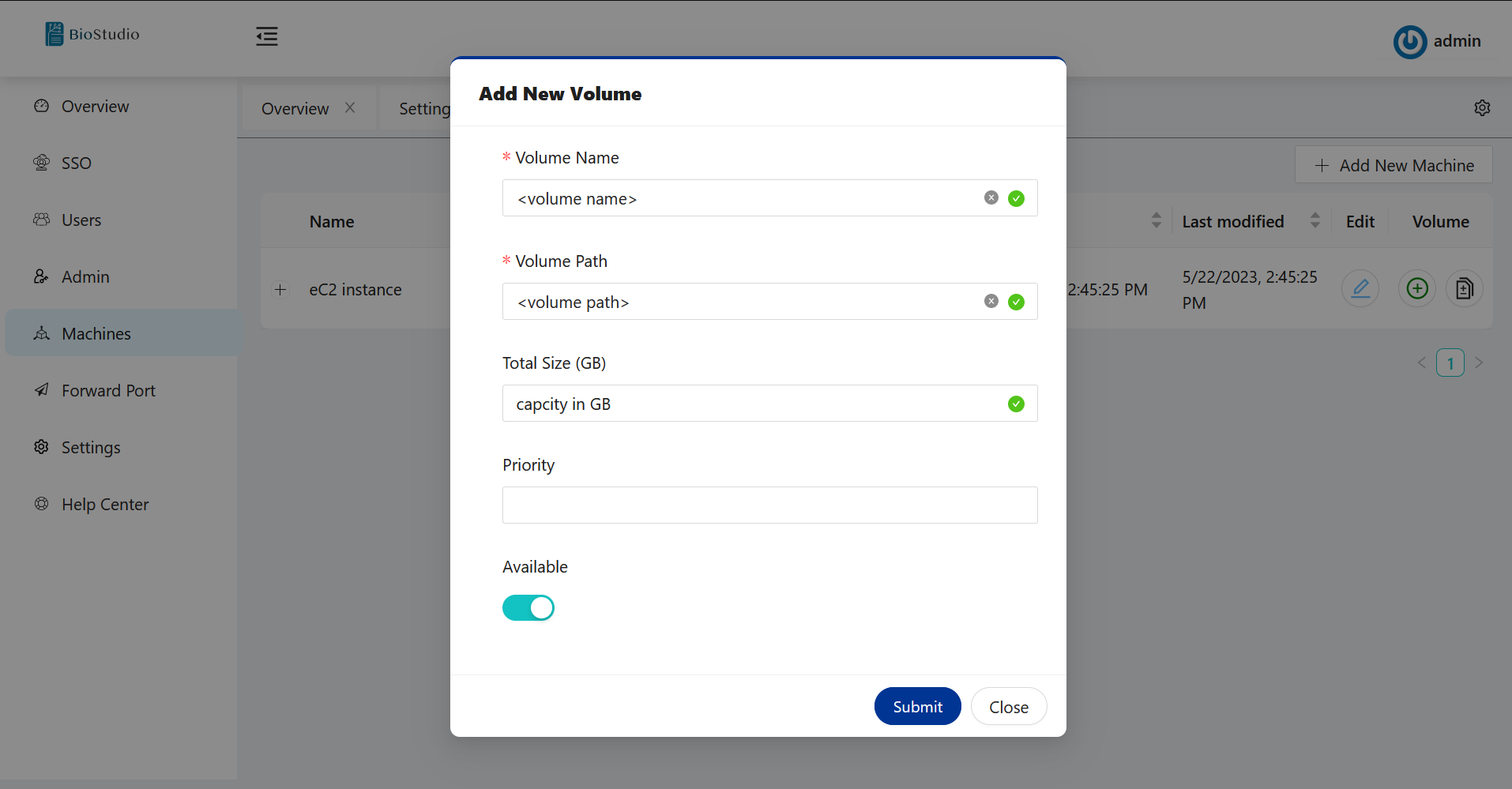
🔶 Please add volume - Update machine Metadata.
# Machines --> Click on + sign to add volume.
# You can use any name for volume, but the volume path will always be /home.
# Workspace is now ready for use.
SSO Set up
¶We are supporting three types of protocol below:
| PROTOCOLS |
|---|
| SAML |
| OPENID |
| OAUTH2 |
🔔 Please contact 📧 support@bioturing.com to add ➕ more protocols and types , If you are not able to find in the list based on your choice. 😊 We are always happy to add more variety of protocols and types.
| SAML : Identity and Access Management |
|---|
| Aliyun IDaaS |
| Keycloak |
| Default |
| OPENID : Identity and Access Management |
|---|
| Default |
| OAUTH2 : Identity and Access Management |
|---|
| Adfs |
| Amazon |
| Apple |
| Auth0 |
| AzureAD |
| BattleNet |
| Bilibili |
| Bitbucket |
| Box |
| Casdoor |
| CloudFoundry |
| Custom |
| Dailymotion |
| Deezer |
| DigitalOcean |
| Discord |
| Douyin |
| Dropbox |
| EveOnline |
| Fitbit |
| Gitea |
| Heroku |
| InfluxCloud |
| Infoflow |
| Intercom |
| Kakao |
| Lastfm |
| Line |
| Mailru |
| Meetup |
| MicrosoftOnline |
| Naver |
| Nextcloud |
| Okta |
| OneDrive |
| Oura |
| Patreon |
| Paypal |
| SalesForce |
| Shopify |
| Slack |
| Soundcloud |
| Spotify |
| Steam |
| Strava |
| Stripe |
| TikTok |
| Tumblr |
| Twitch |
| Typetalk |
| Uber |
| VK |
| Wepay |
| Xero |
| Yahoo |
| Yammer |
| Yandex |
| Zoom |
SSO configuration
¶🔆 Login to Dashboard using admin credential.
🔗 https://<Domain Name>/dashboard
🔗 http://<Domain Name>/dashboard
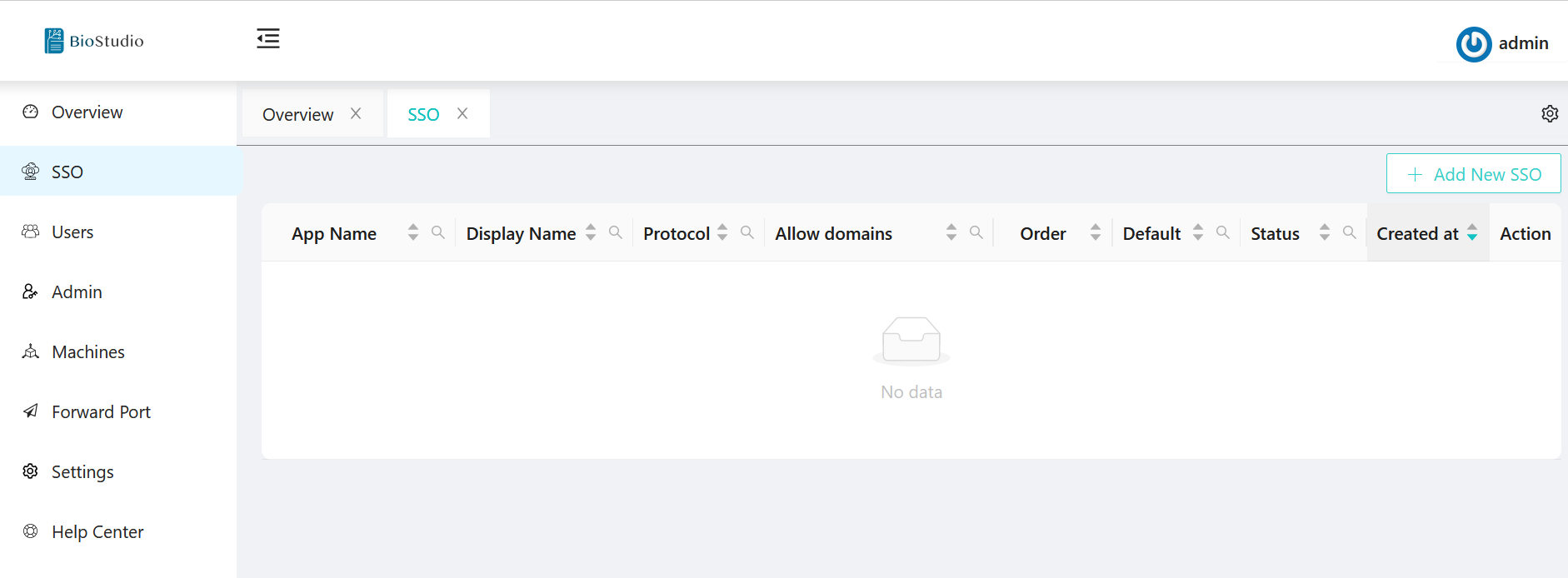
🔆 Select SSO on ⬅️ left side menu.
🔆 Click on ➡️ + Add New SSO push button.
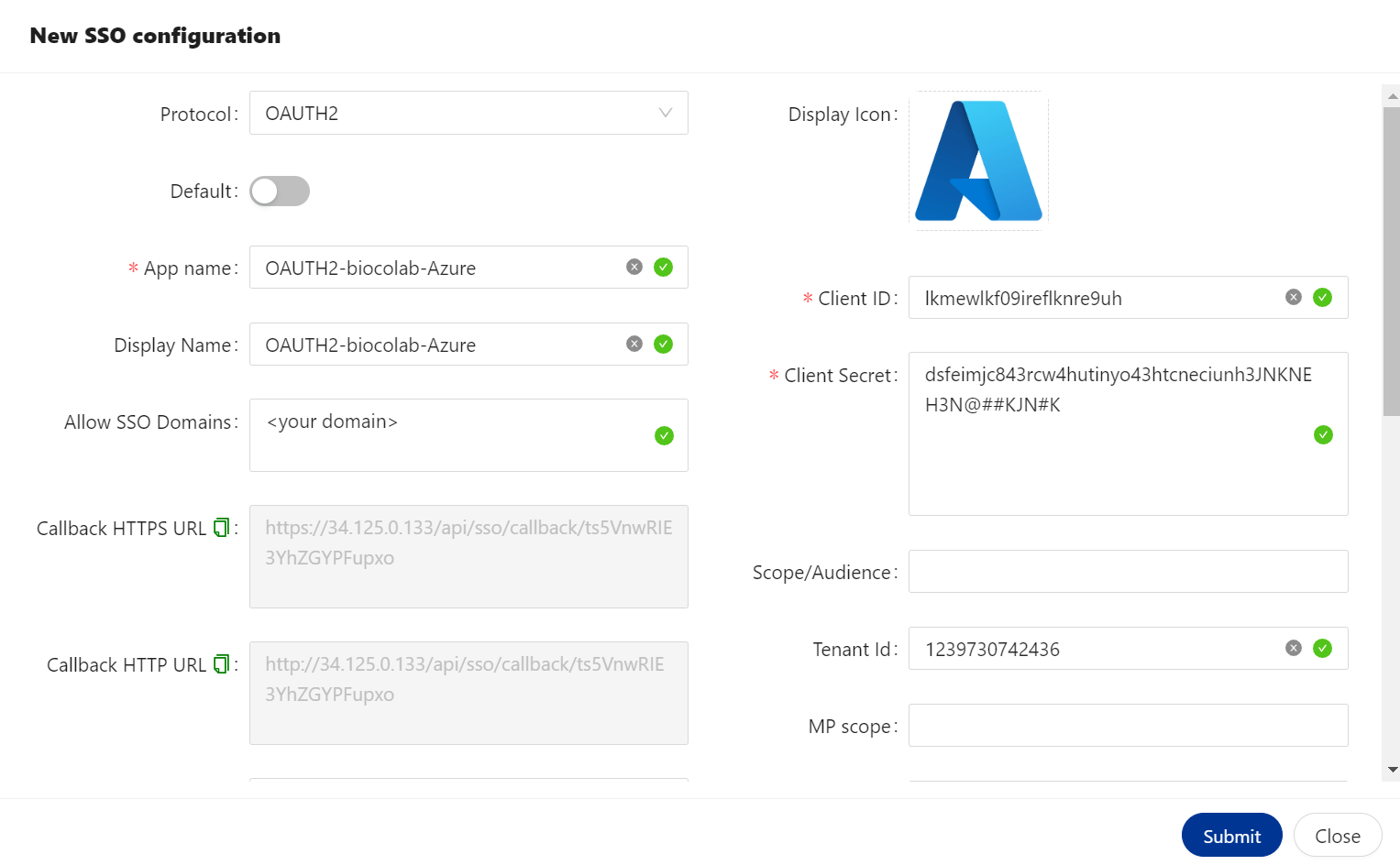
🔶 New SSO configuration.
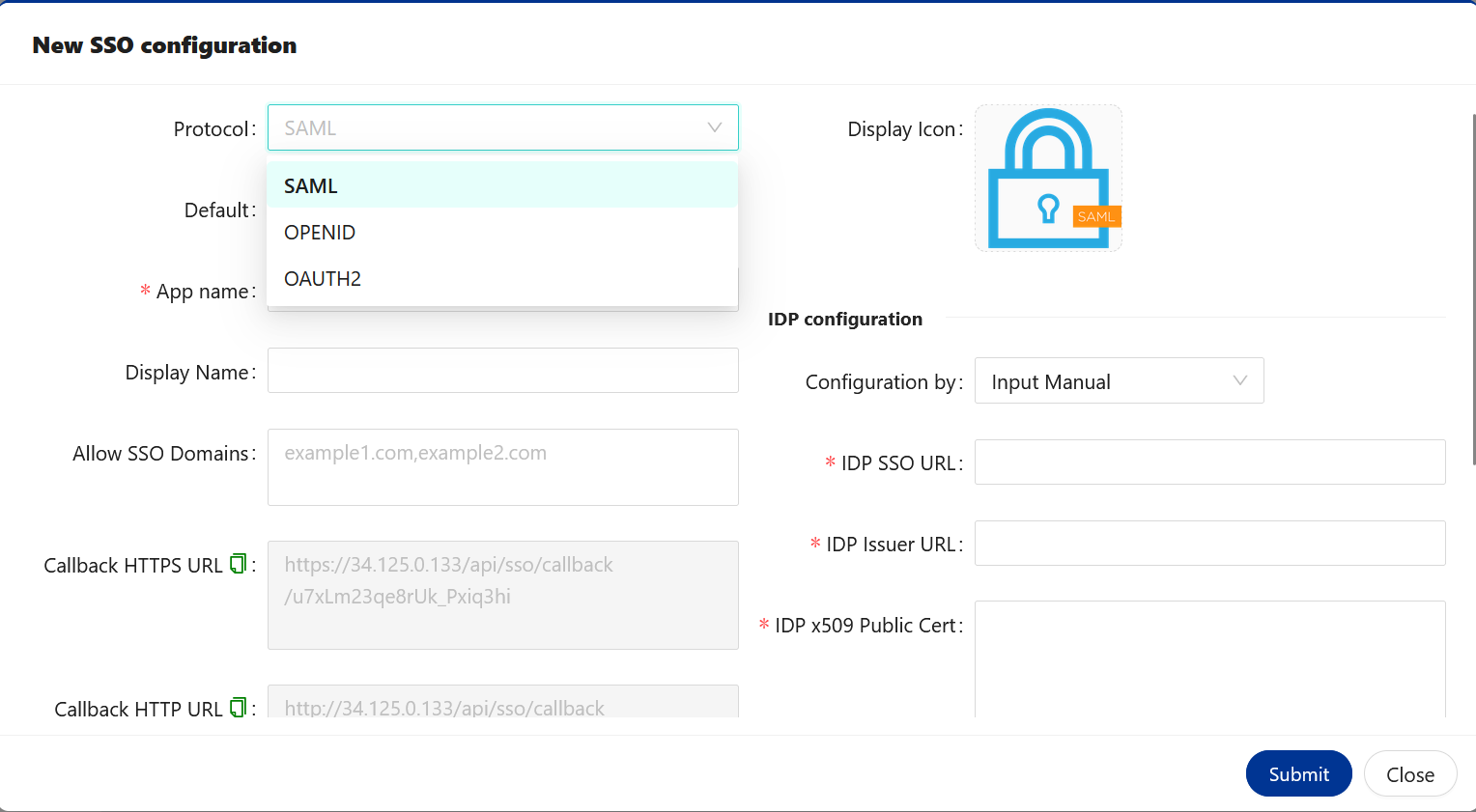
| Option | Description |
|---|---|
| Protocol | Rules that you would like to select for SSO configuration. It could be SAML, OPENID or OAUTH2. |
| Default | If you are configuring - multiple SSO service providers. You can mark one of them to use as a default by enabling Default option. |
| App name | Specified application name. You can provide any name. |
| Display Name | Name that you would like to use for SSO configuration. You can provide any name. |
| Allow SSO Domains | This option allow you to restrict access to an application. Only selected domain are allow to access this application. Kindly provide your domain name, if you wish to allow to access user’s belongs to your domain. |
| Callback HTTPS URL | We must use this URL to configure SSO, if BioStudio configured with SSL access. So that once Authentication Successful. User will allow to access BioStudio application. |
| Callback HTTP URL | We must use this URL to configure SSO, If BioStudio configured without SSL. So that once Authentication Successful. User will allow to access BioStudio application. |
| Type | Select what type of IDP service provider you are going to use. value will be changed based on selected Protocol. |
| Subtype | If you have more type of IDP. Type vlaue in Subtype. |
| Disable SSL | By default SSL is disabled. If you would like to auth. with server’s keys. Enable it and you can have public and private key to configure SSO. |
| Configuration by | You can select to provide value by manual, Ready from XML content or Ready from XML file URL. |
| IDP SSO URL | Identity Provider SSO URL that you can get it from IDP. |
| IDP Issuer URL | Identity Provider Issuer URL that you can get it from IDP. |
| IDP x509 Public Cert | Identity Provider certificate. That you can download or copy the content and provide it here. |
🔔 Note: Callback HTTPS URL, Callback HTTP URL is not static and will change each time for new SSO configuration for the same potocol.
🔐 Solution: We can provide dummy values to mendatory fields and click on Submit button to 💾 save. Once saved, We can provide Callback URL to SSO IT department for further configuration. Collect all required values after configuring Callback URL from IDP and configure it further by update SSO.
🔶 SAML Protocol
SAML is an XML-based authentication protocol in which Identity Providers (IdP) -- entities that manage and store user credentials -- exchange digitally signed XML documents (SAML Assertions) allowing an end-user to access a **Service Provider **(SP).
Here IDP could be Aliyum IDaas, KeyCloak, auth0, or any of IDP service provider, which come under default.
BioStudio is comes under service provider.
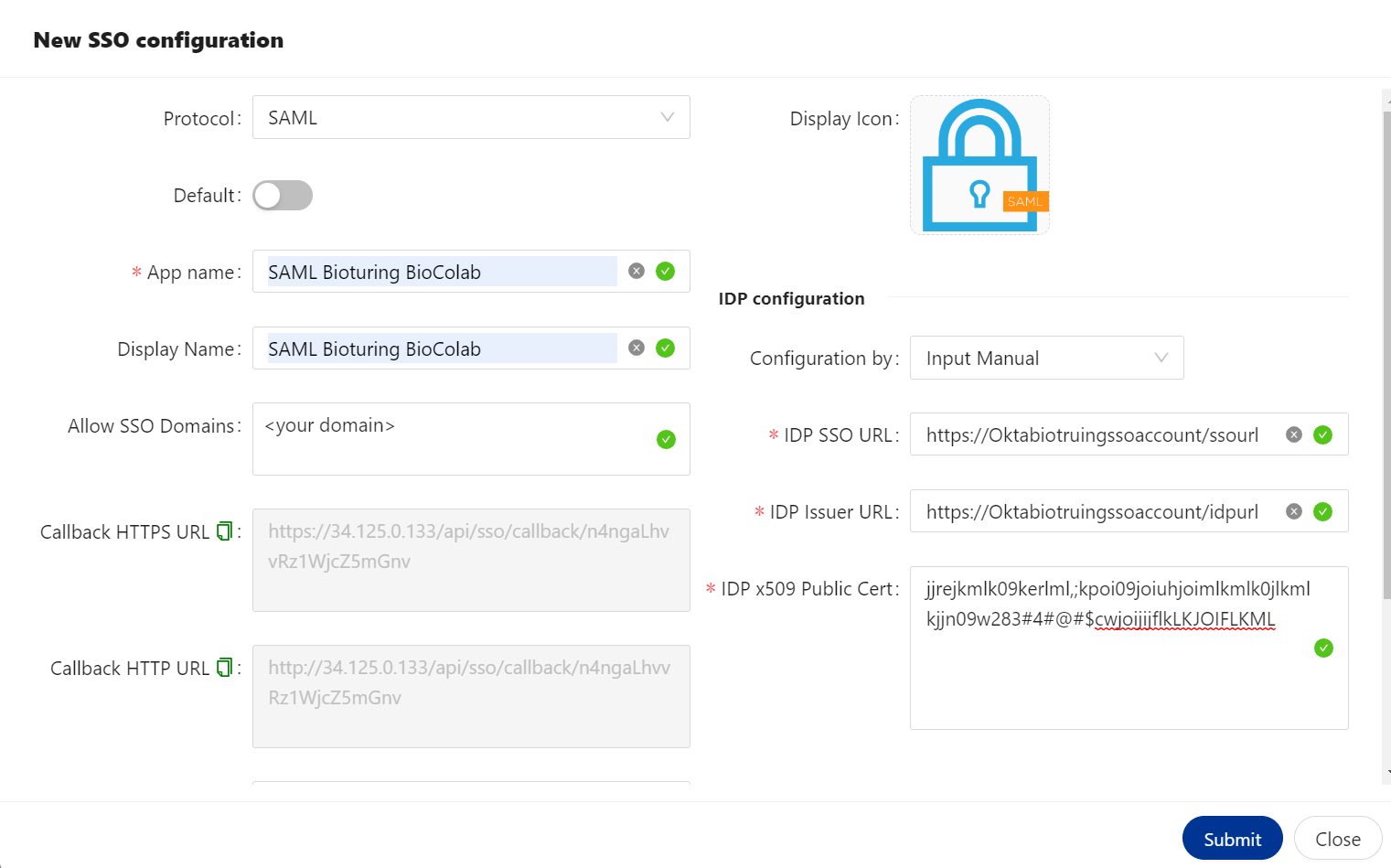
🔷 SAML Protocol Configured.
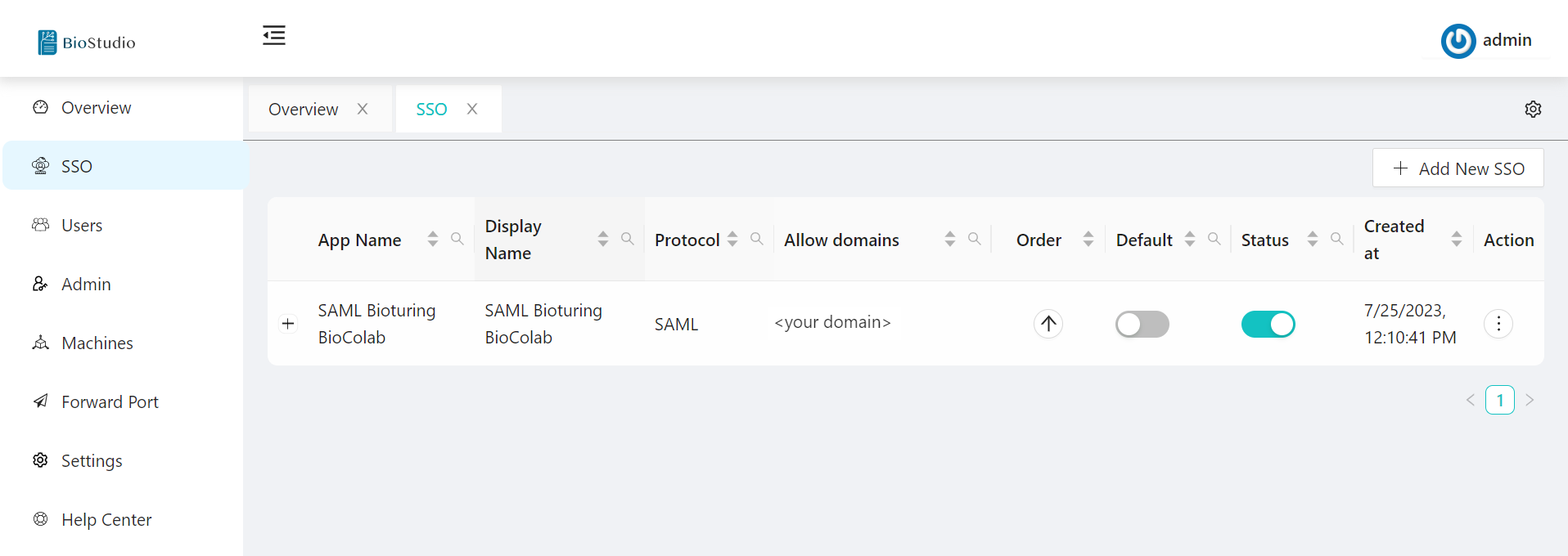
🔷 SAML Protocol view metadata.
# Once clicked on Submit button. You can view metadata by clicking + ( Plus sign)
🔷 SAML metadata.
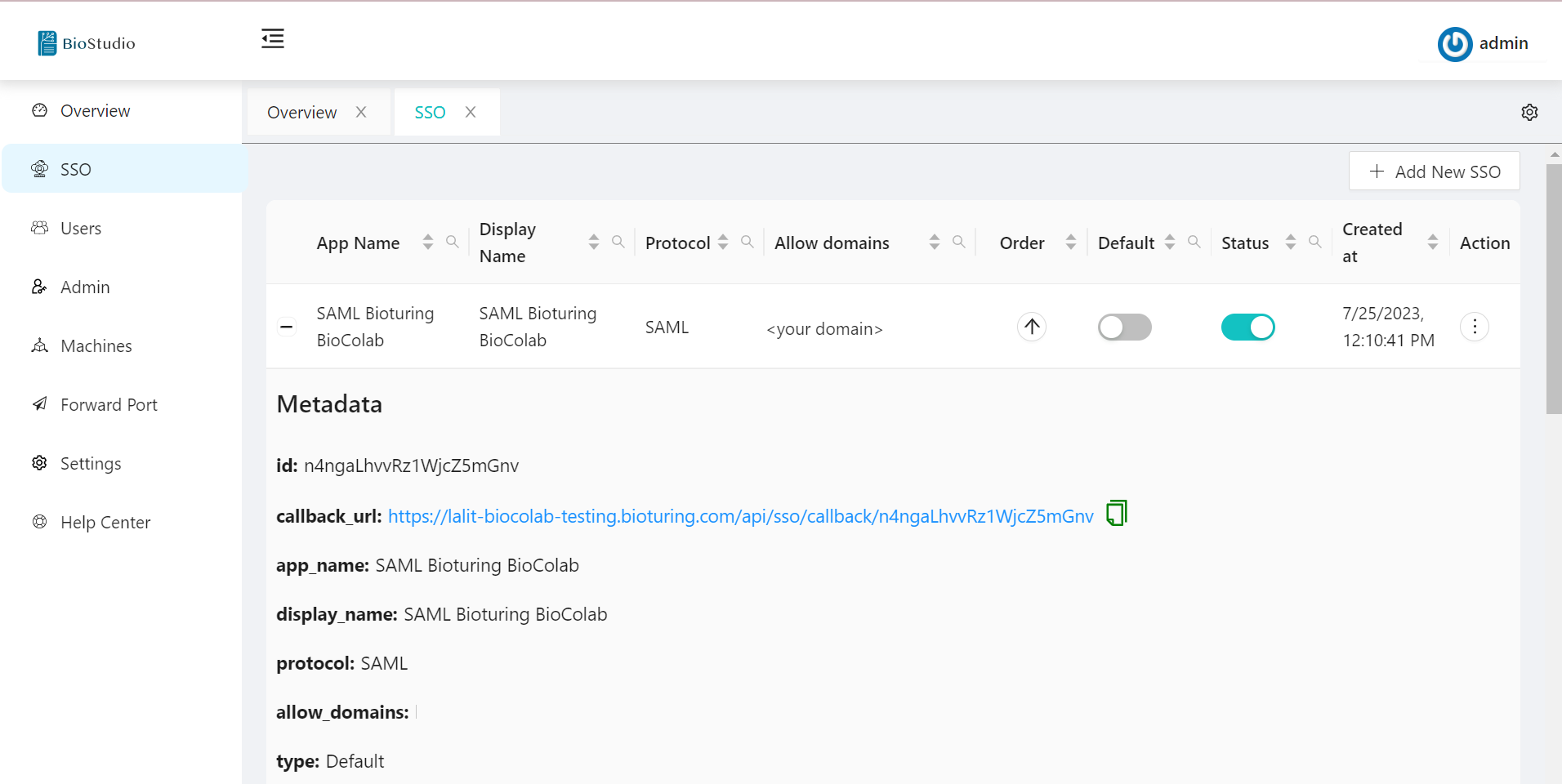
🔷 SAML Protocol update / delete method.
You can update the values of an existing SSO configuration whenever you want by selecting the update option for SSO.
# In Order to update or delete existing SSO configuration. Please follow steps are given below.
1. Login to the dashboard using Admin credential.
2. Select SSO option available on left side menu.
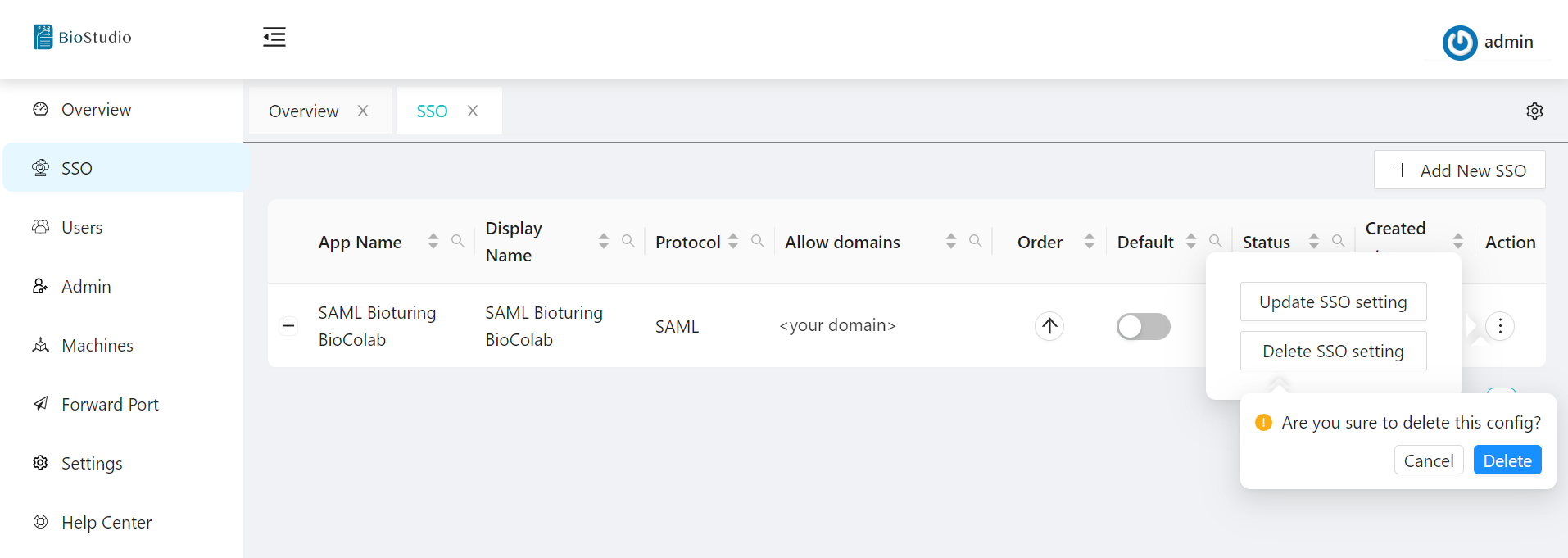
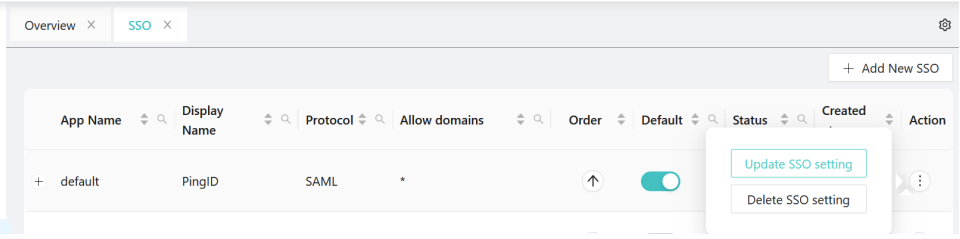
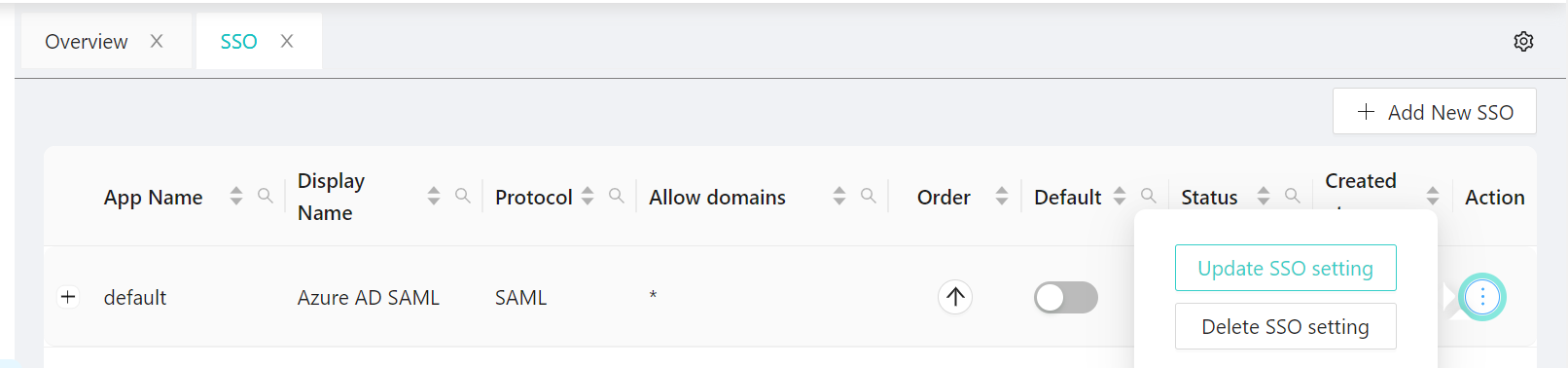
3. Select SSO Application -- Action three DOTS to select your choice to update or delete.
# option - Action.
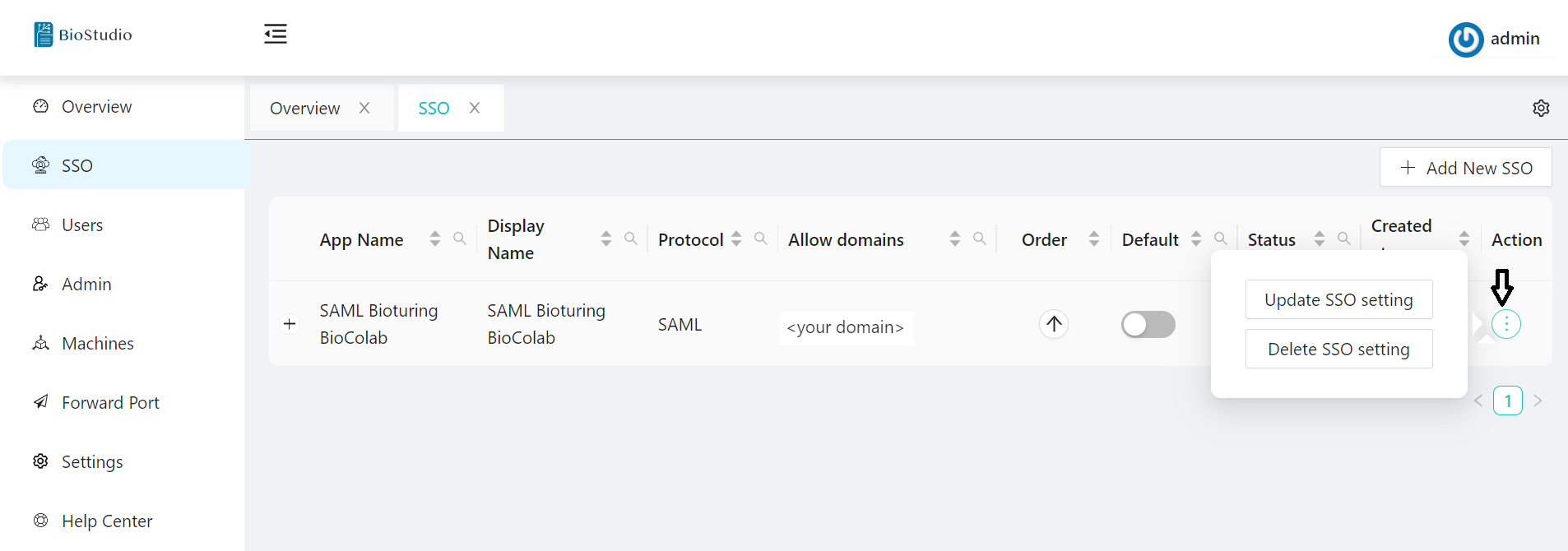
# option - Update.
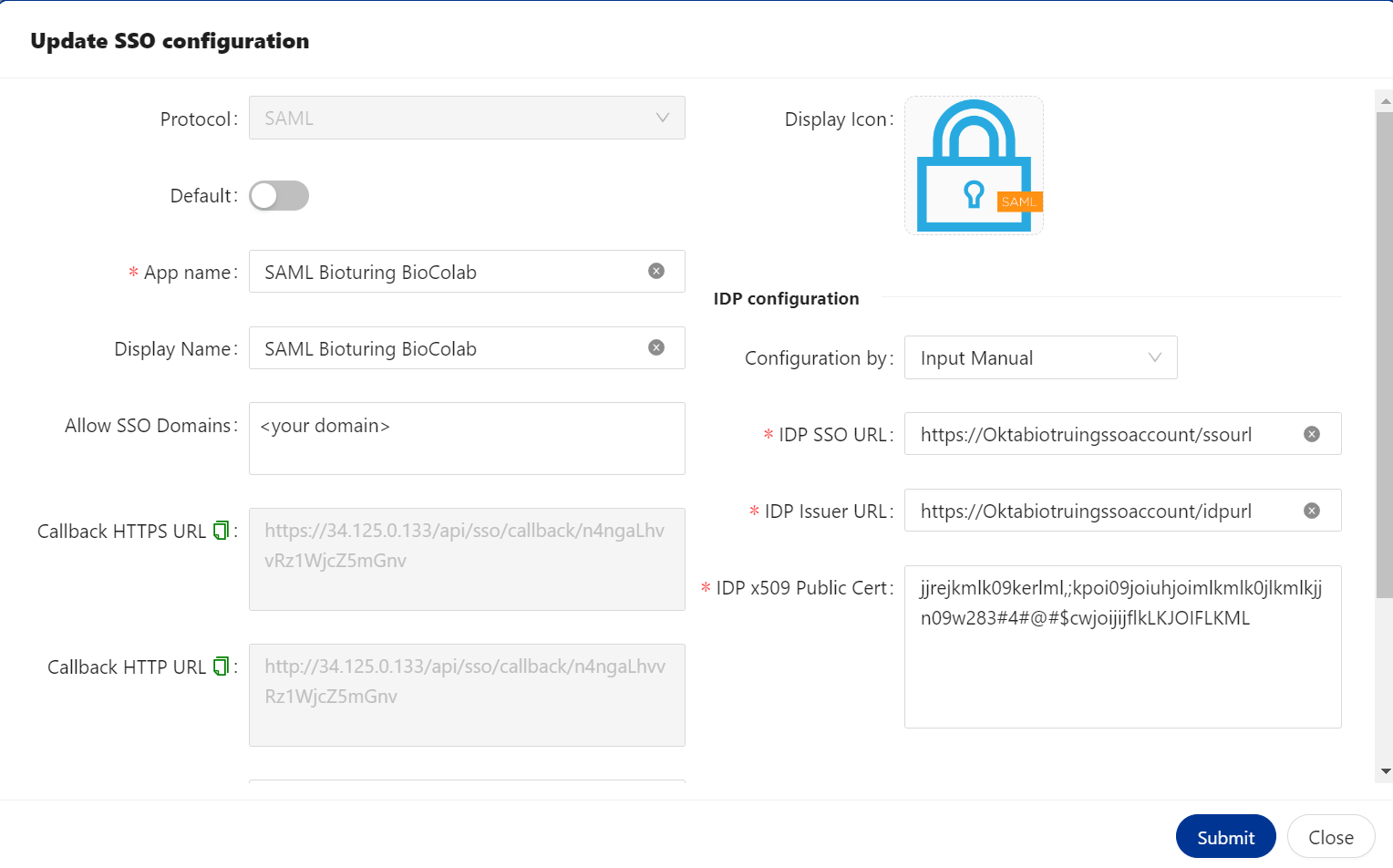
# option - Delete.
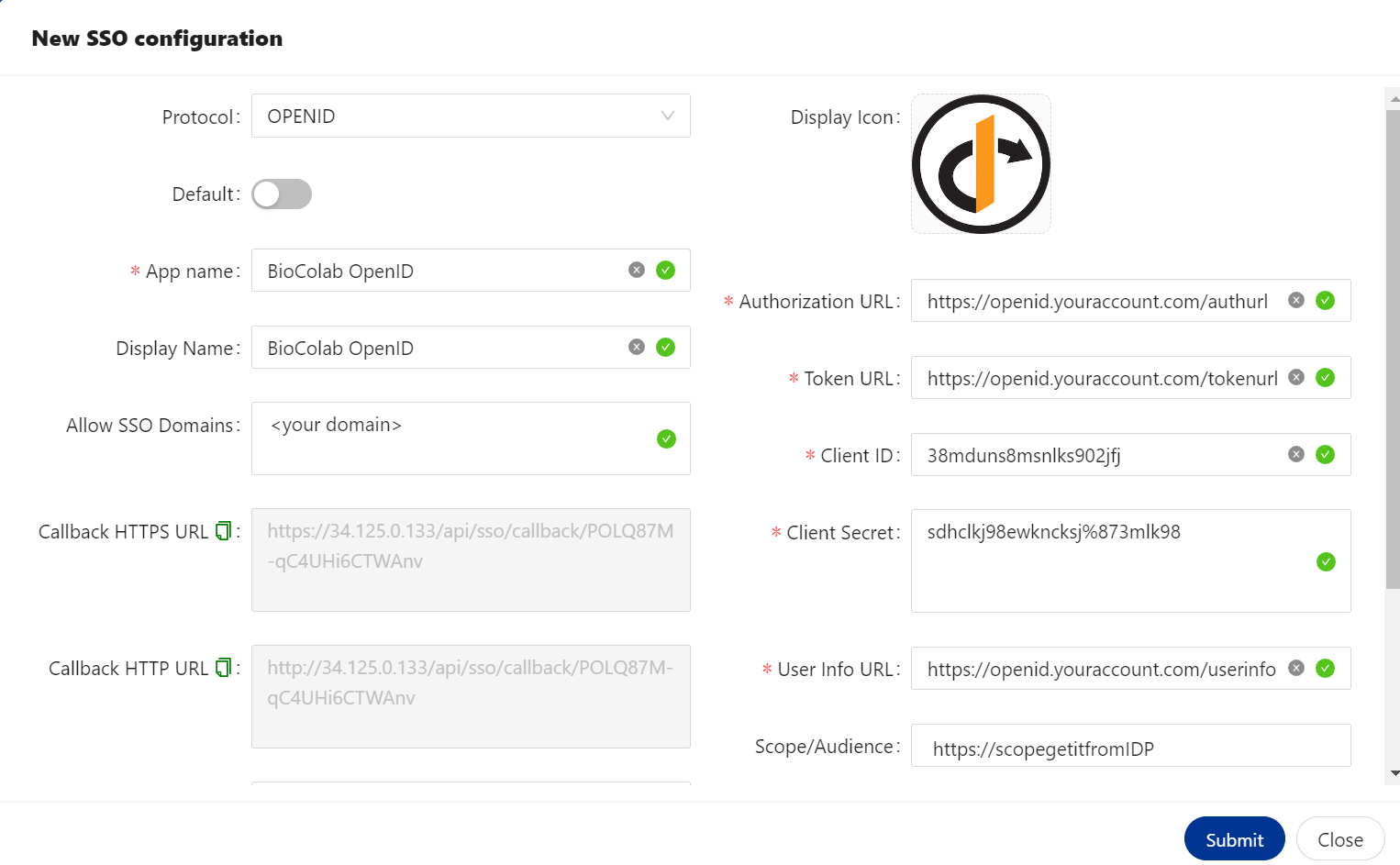
🔶 OPENID Protocol.
OpenID Connect is an interoperable authentication protocol based on the OAuth 2.0 framework of specifications (IETF RFC 6749 and 6750). It simplifies the way to verify the identity of users based on the authentication performed by an Authorization Server and to obtain user profile information in an interoperable and REST-like manner.
🔷 OPENID configured.
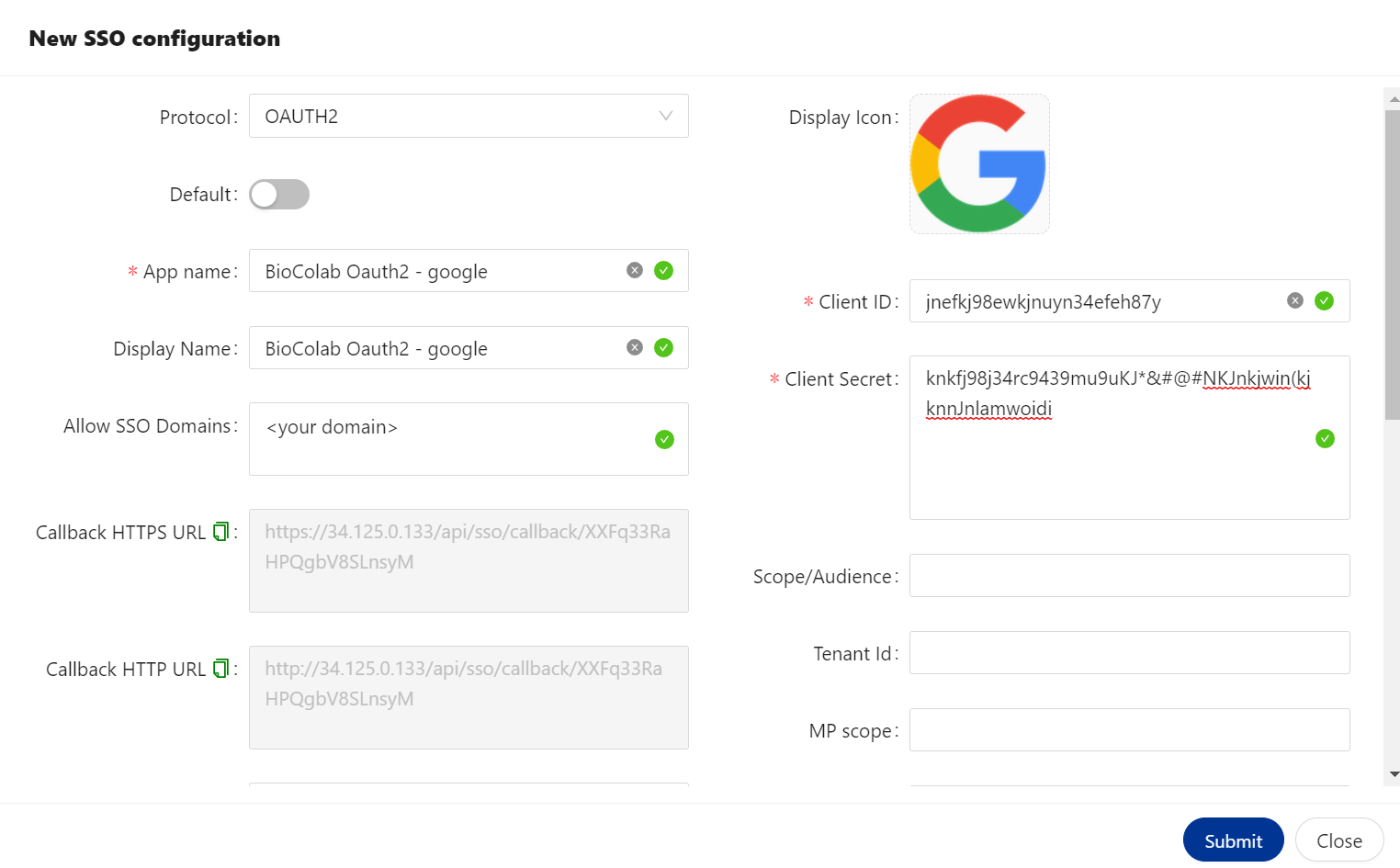
🔶 OAUTH2 Protocol.
The OAuth 2.0 is the industry protocol for authorization. It allows a user to grant limited access to its protected resources. Designed to work specifically with Hypertext Transfer Protocol (HTTP), OAuth separates the role of the client from the resource owner. The client requests access to the resources controlled by the resource owner and hosted by the resource server. The resource server issues access tokens with the approval of the resource owner. The client uses the access tokens to access the protected resources hosted by the resource server.
We are providing all type of IDP’s to configure SSO using OAUTH2. List is given above.
🔷 OAUTH2 configured with Google.
🔷 OAUTH2 configured with Azure.
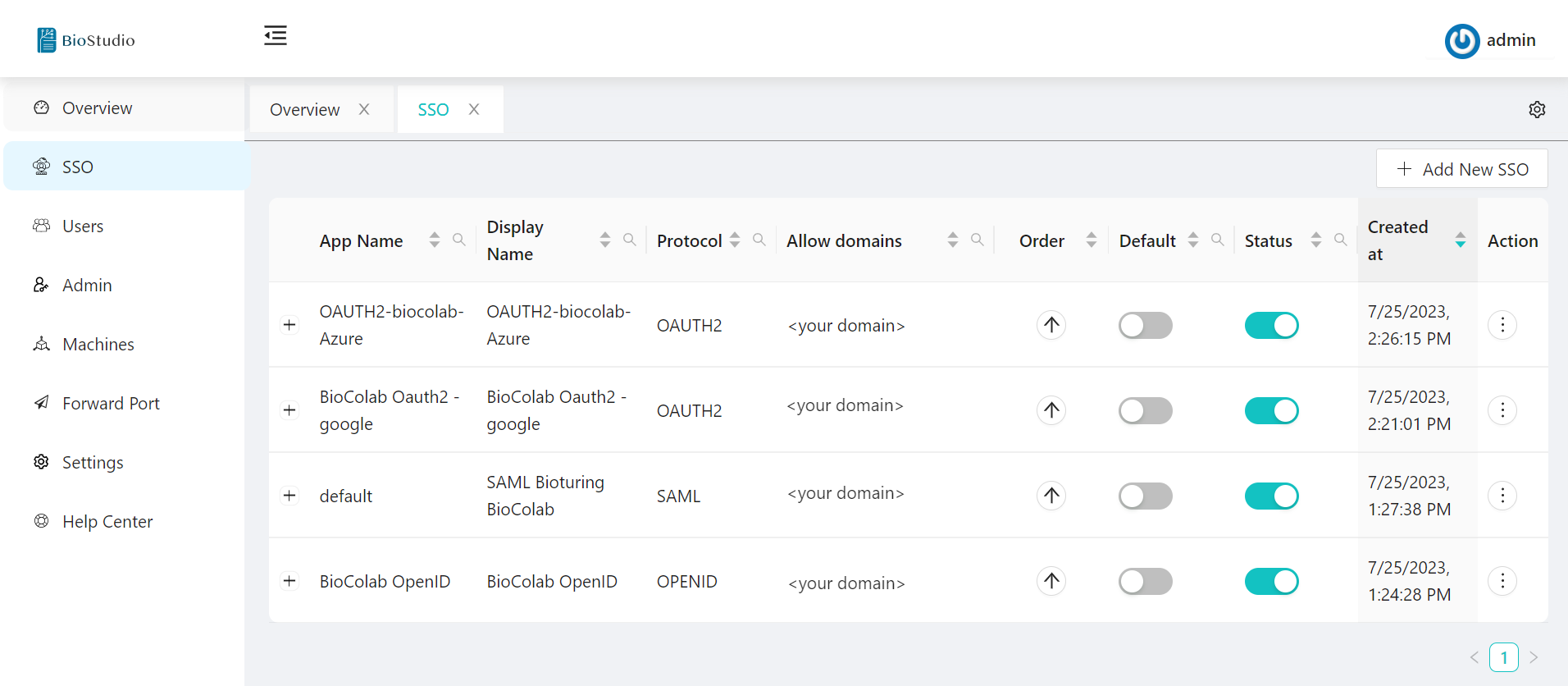
In above explanation, We configured four types of SSO. It will appears to users during login to decide login method.
SSO set up with Okta ¶
⭕ Login to Okta Account for SAML / OPENID.
⭕ We are illustrating for SAML.
⭕ Click on admin account push button and login to your Okta admin account.
NOTE: Kindly follow the instructions and steps based on your infrastructure / IT department team.
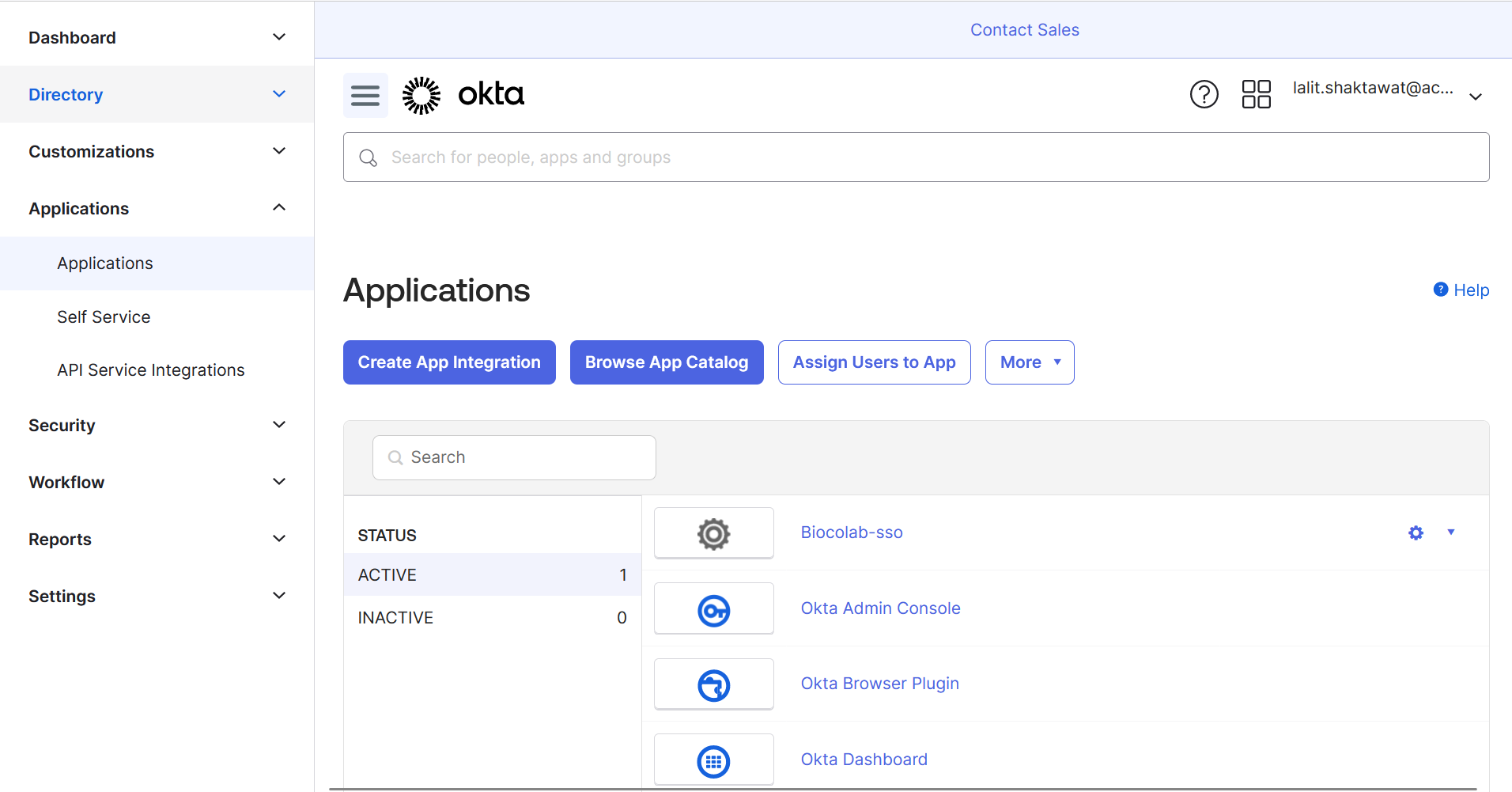
🔶 Click on Create App Integration.
🔶 Select SAML 2.0
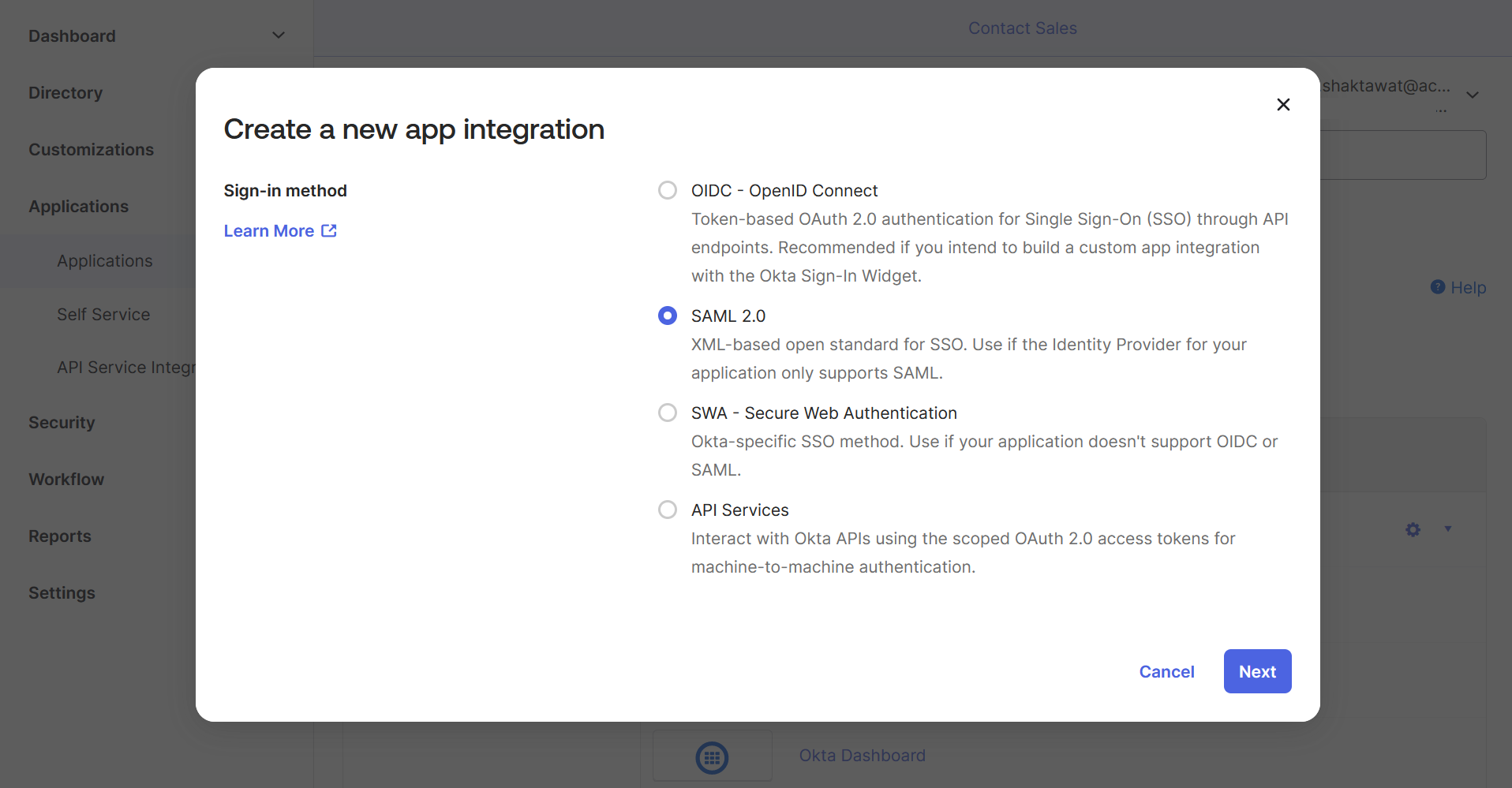
🔶 Write App name
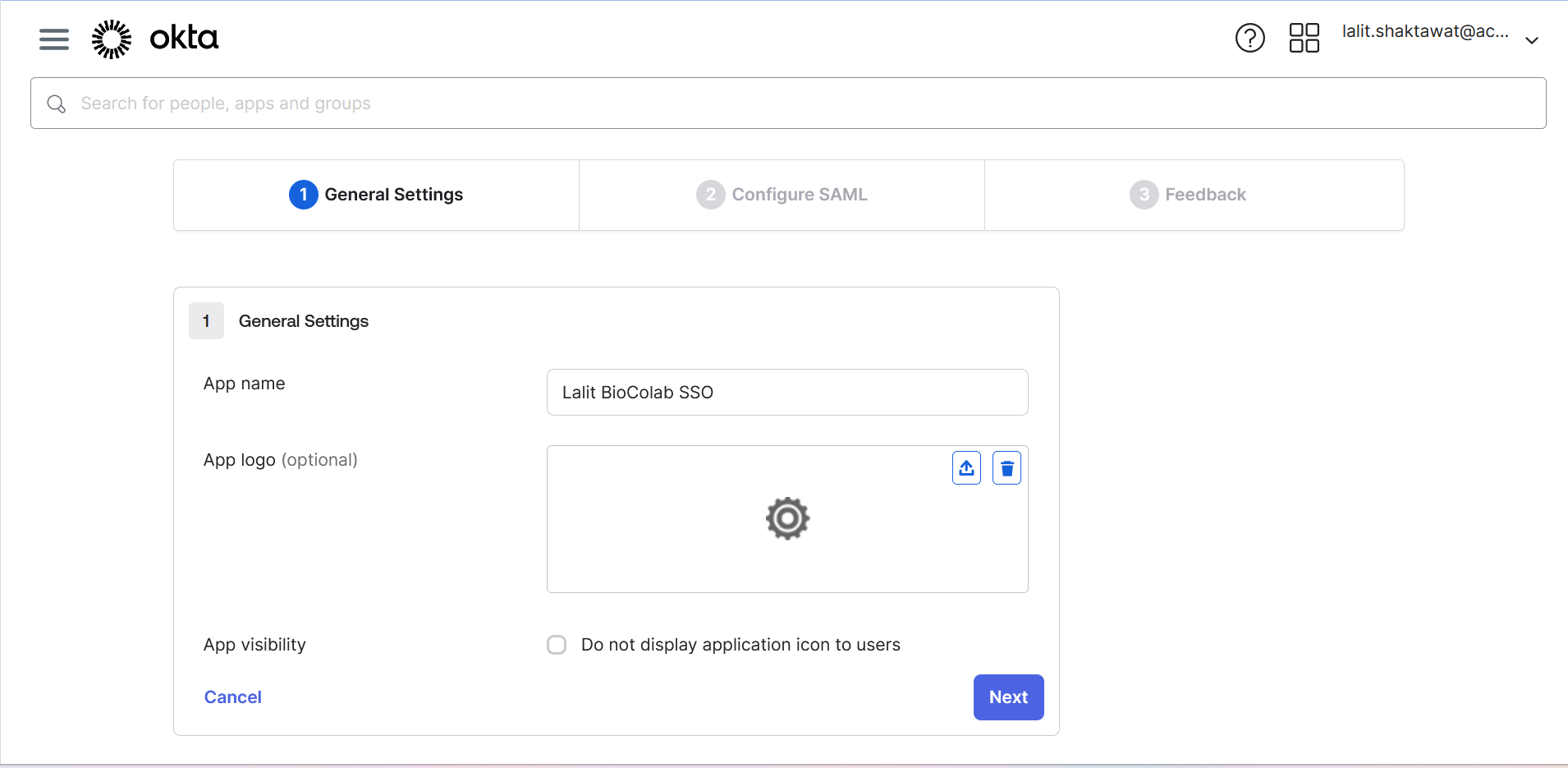
🔶 Upload application logo ( If you wish ).
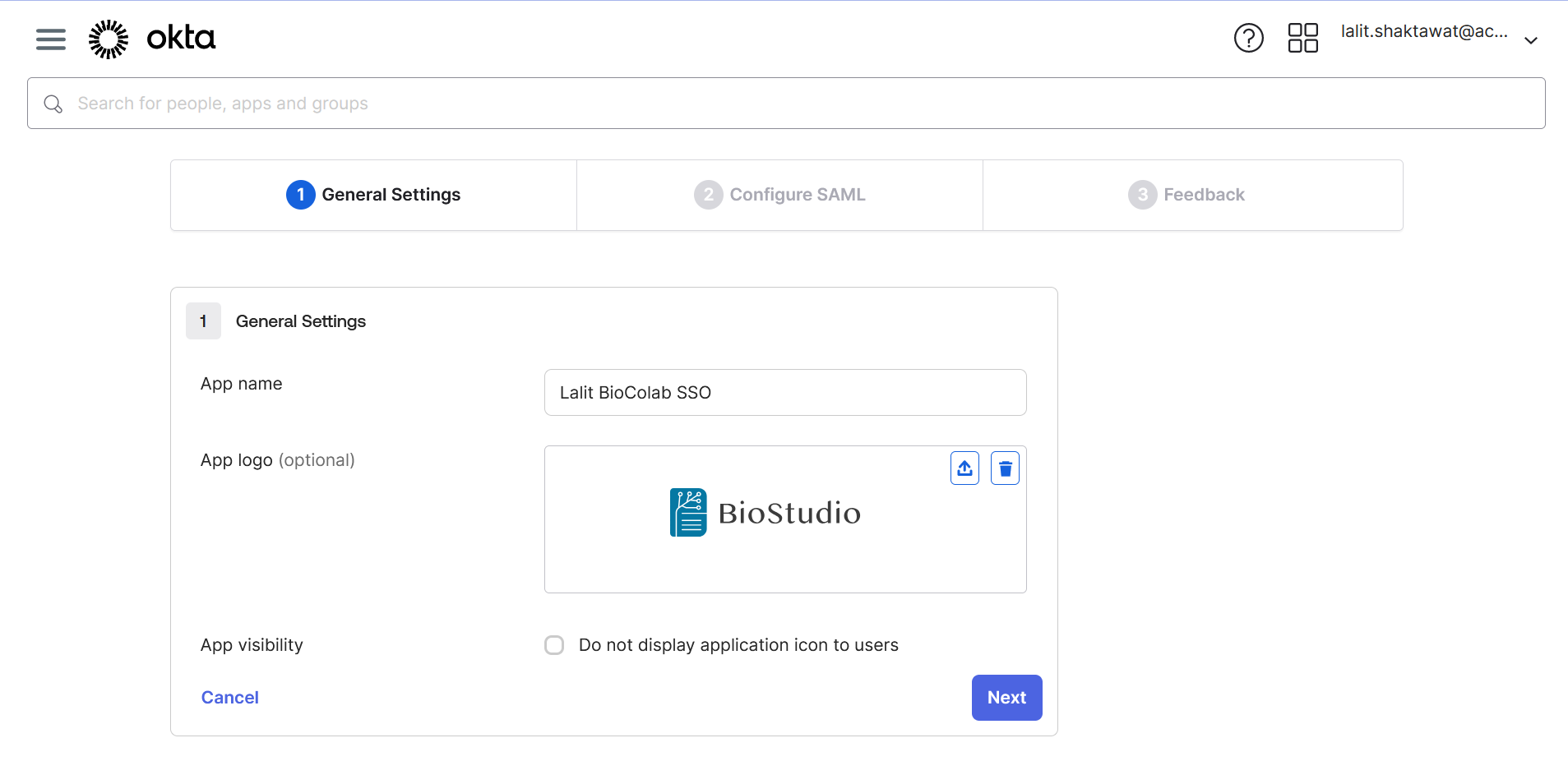
🔶 On BioStudio dashboard login. Select SSO ➡️ Add New SSO.
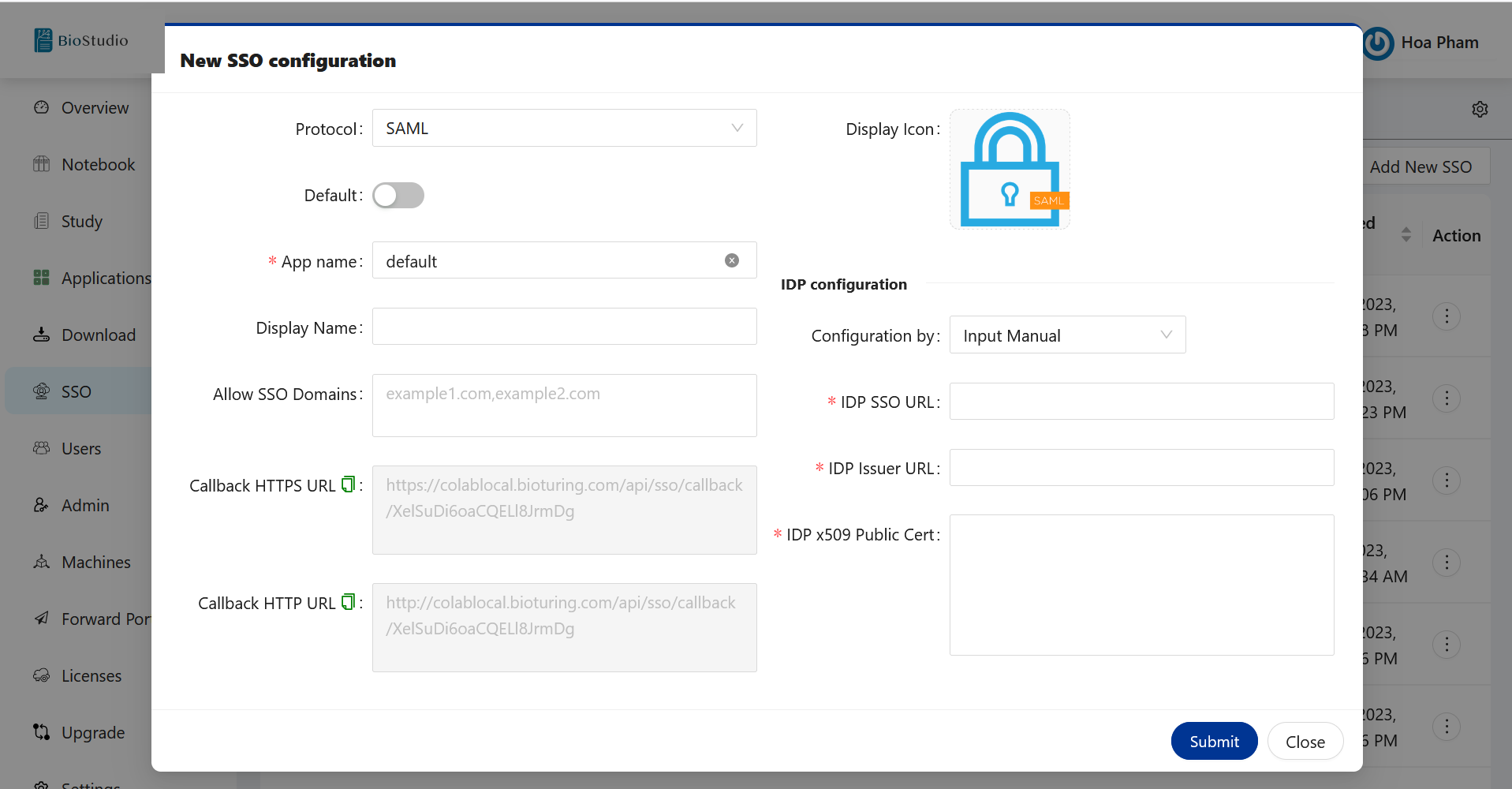
🔶 Copy callback URL from New SSO login and update on Okta.
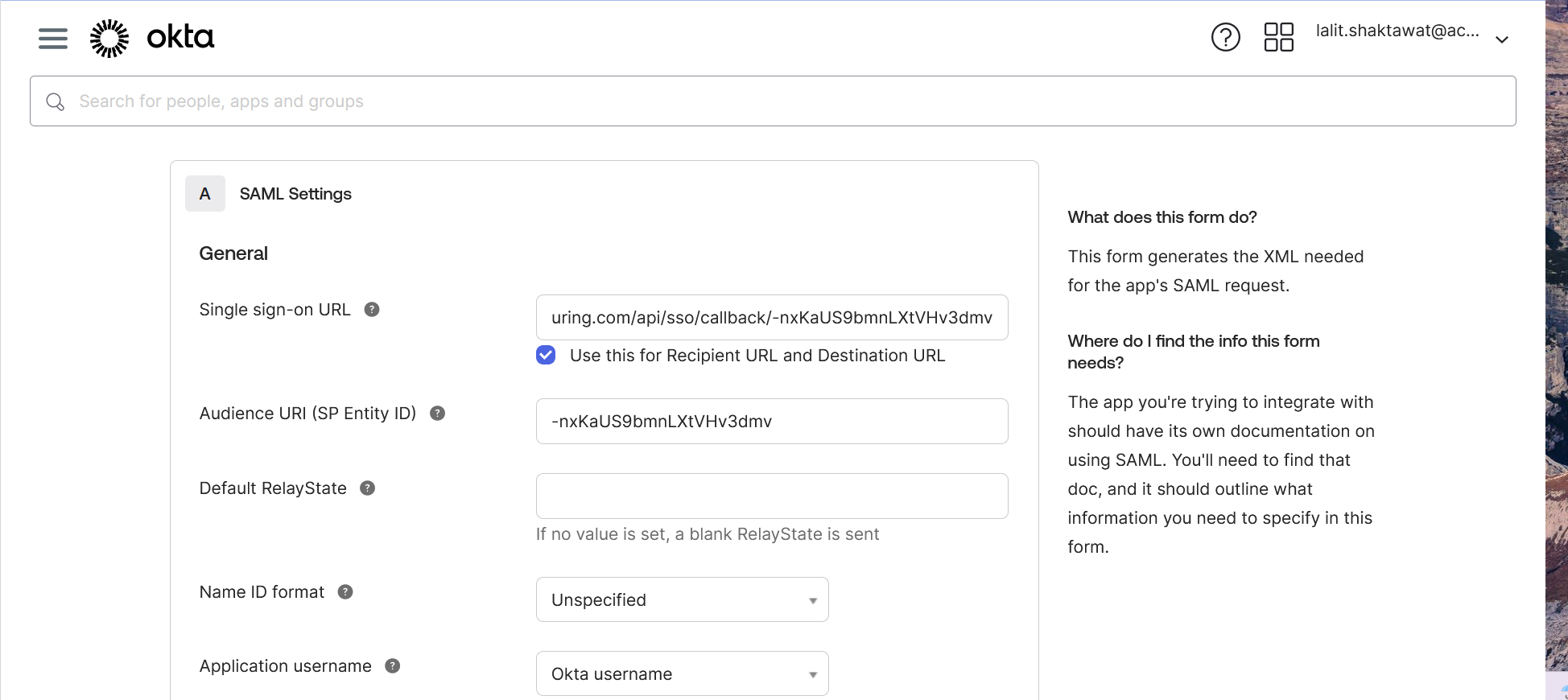
🔶 Scroll down and click on Next
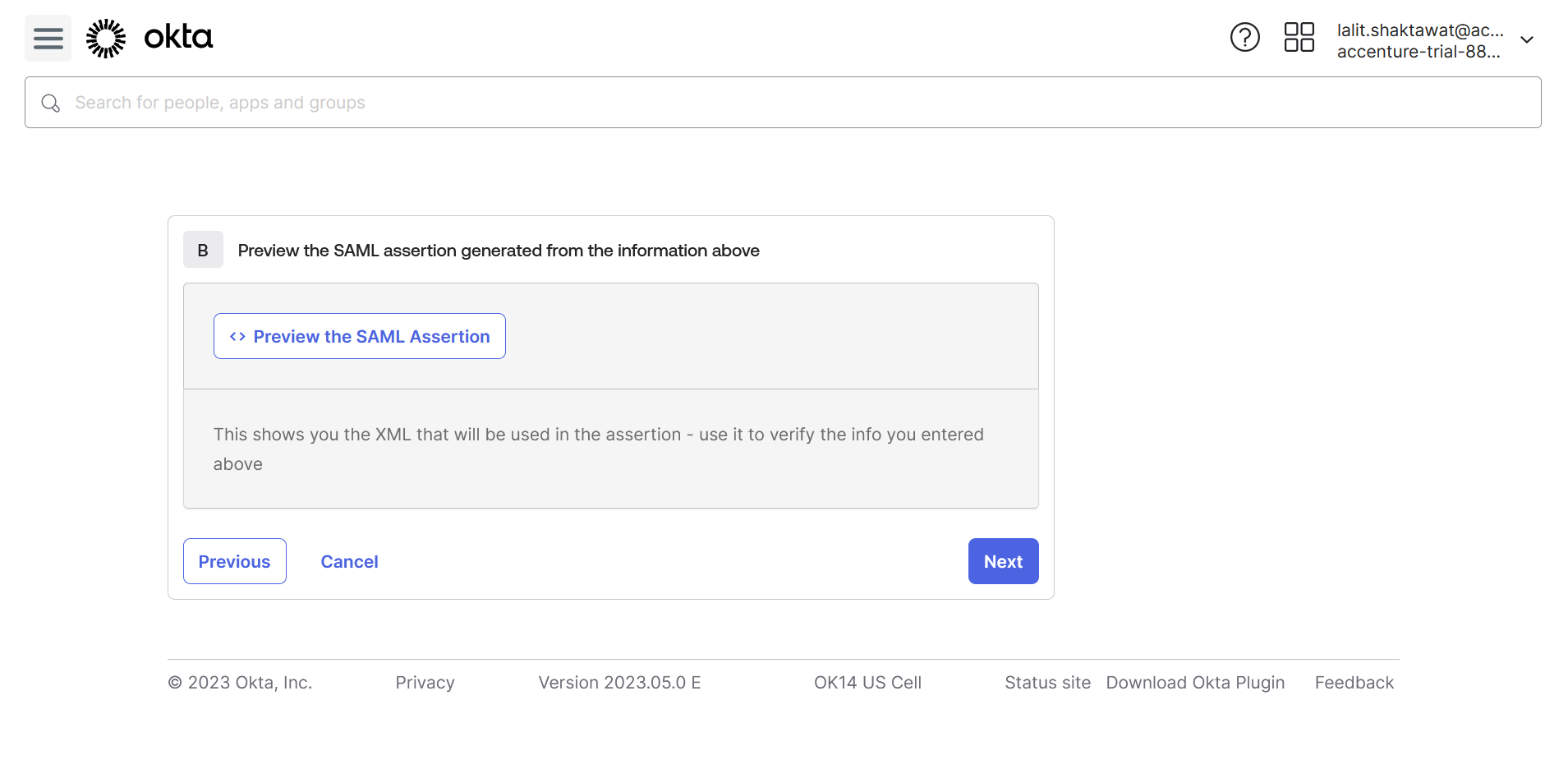
🔶 Follow Step3: I’m an Okta customer adding an internal app.
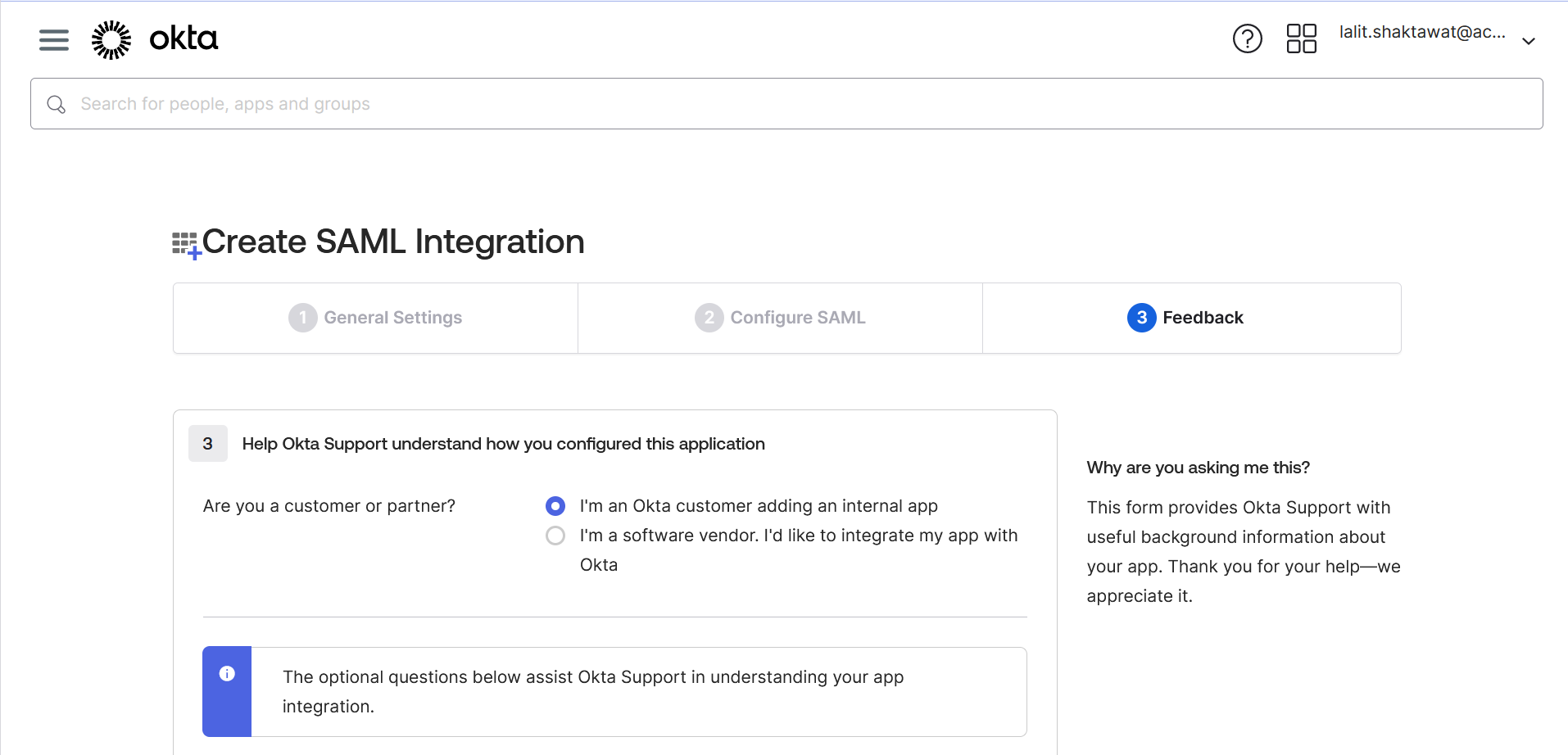
🔶 Click on Finish.
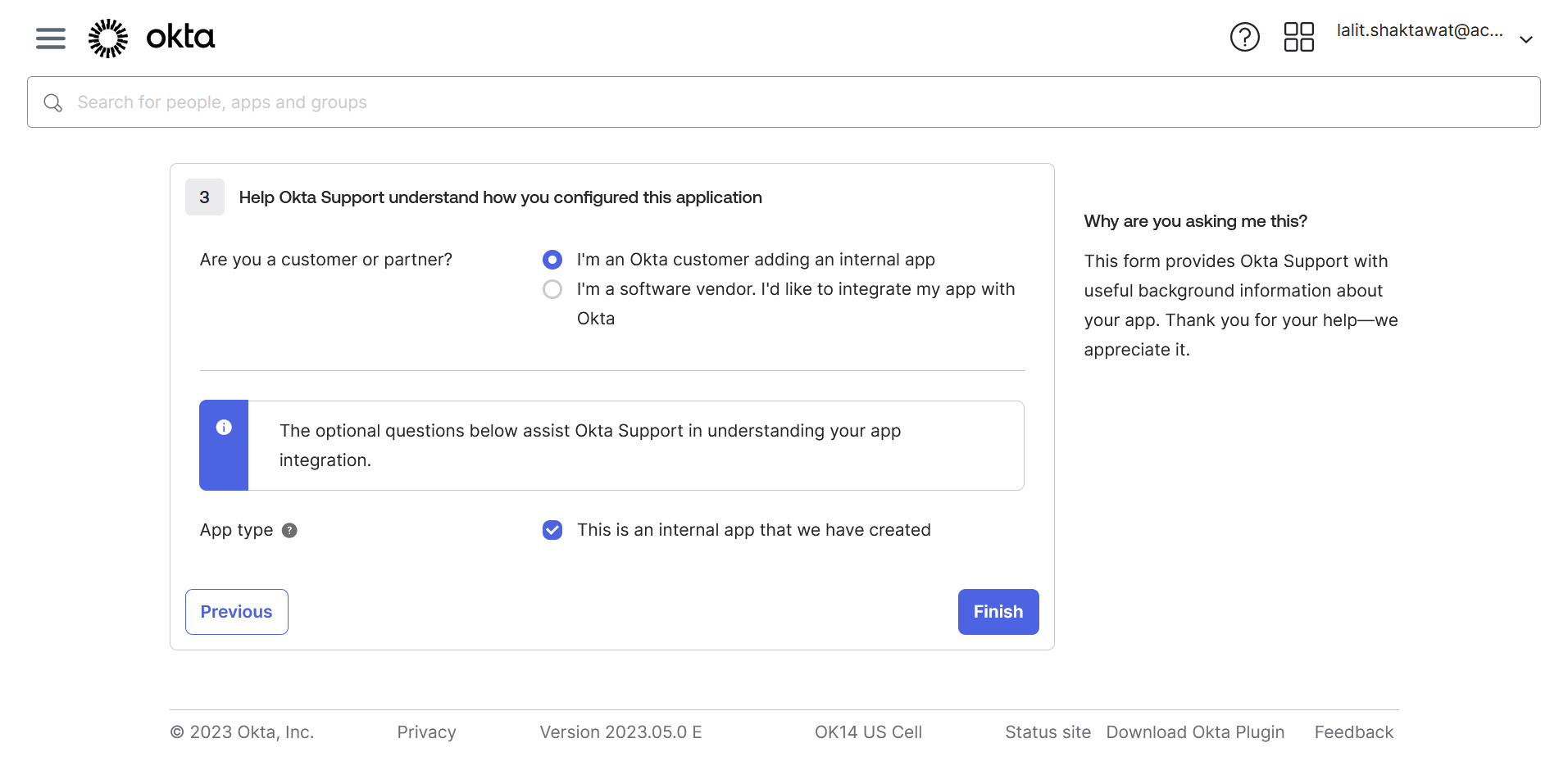
🔶 SAML config data is ready.
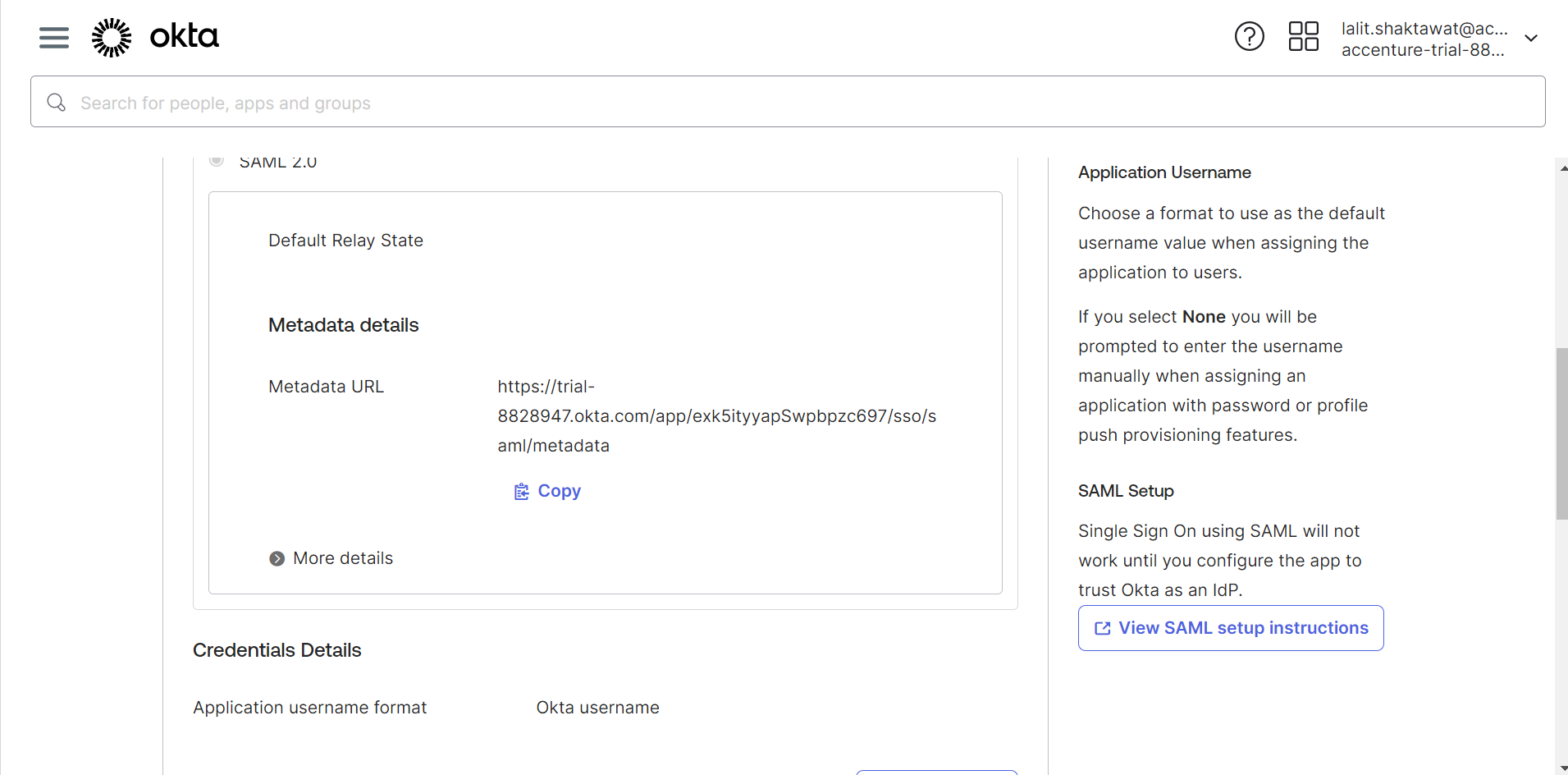
🔶 View SAML setup instructions.
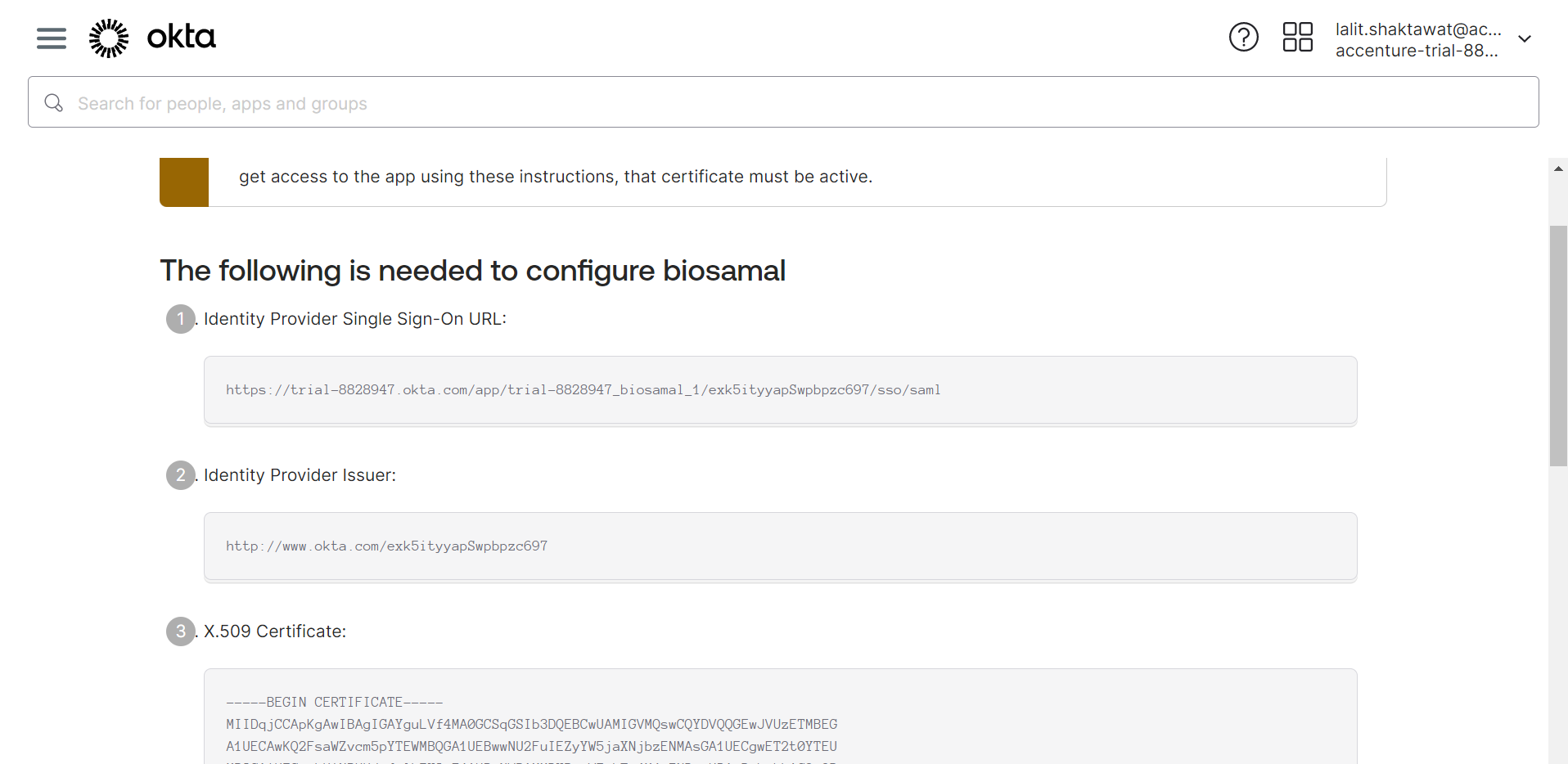
🔶 Get the value from Okta and fill on SSO BioStudio. Click on Submit.
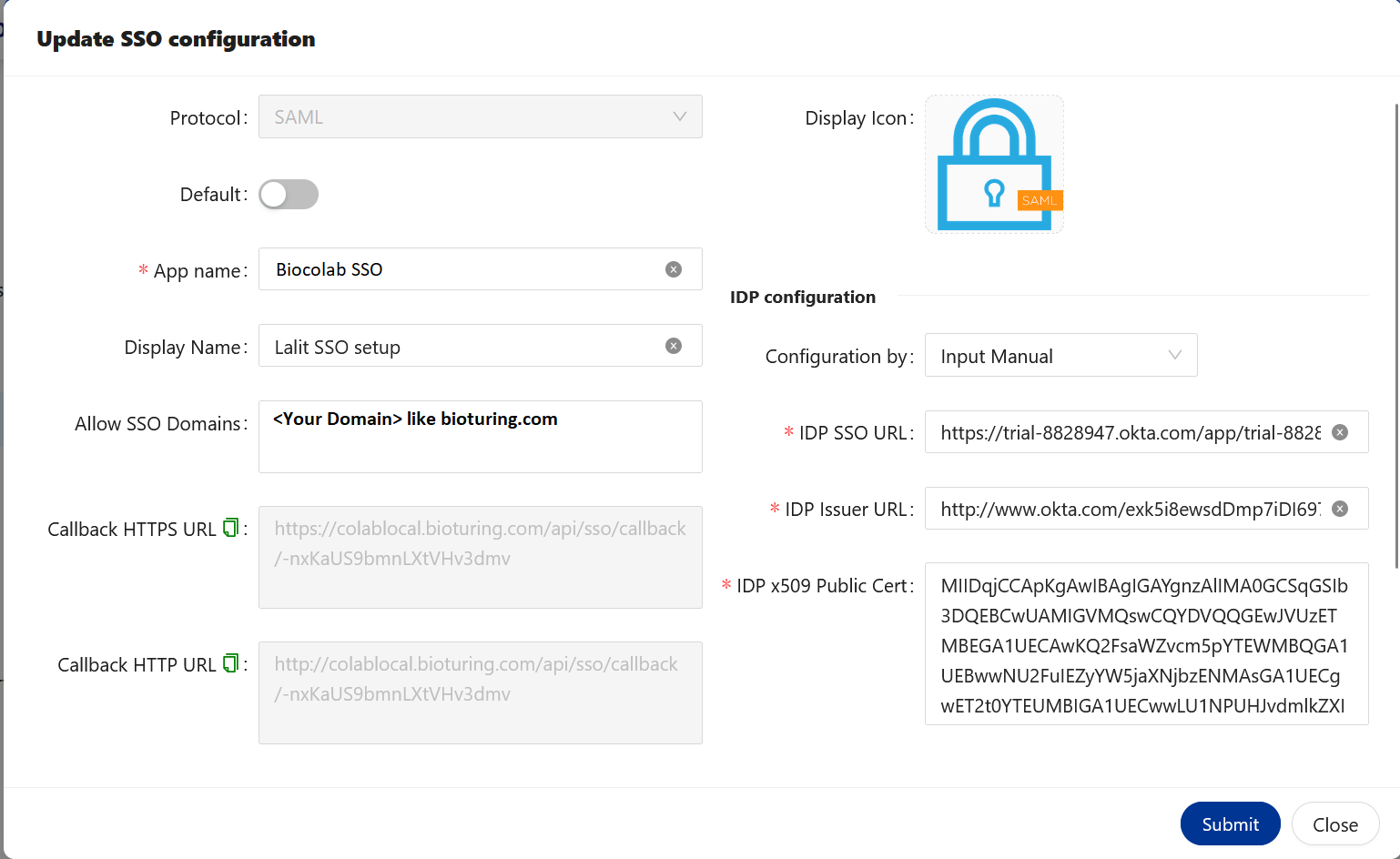
🔶 User assignment to Application.
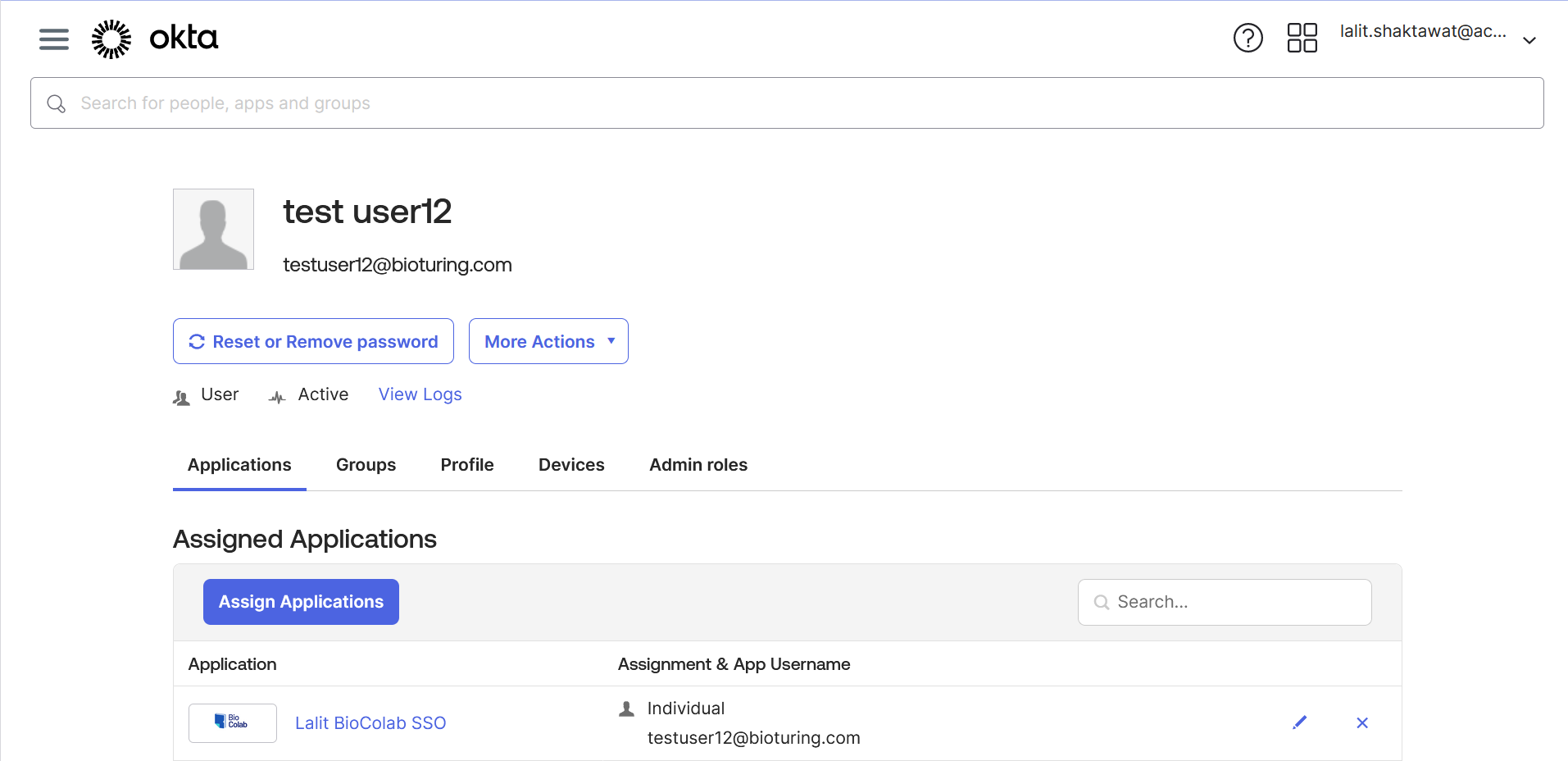
🔶 Assign User to Application.
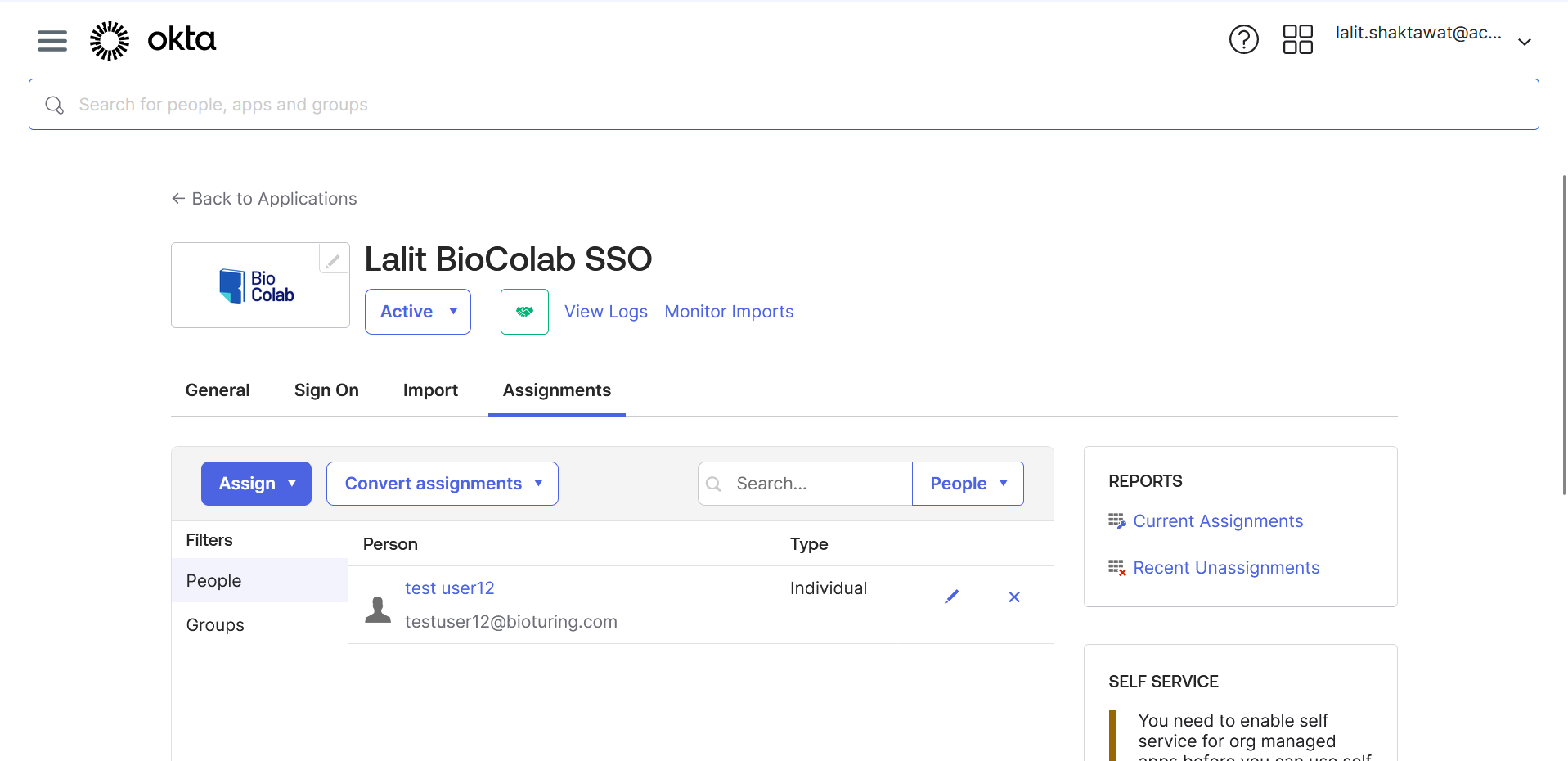
🔶 Login to BioStudio.
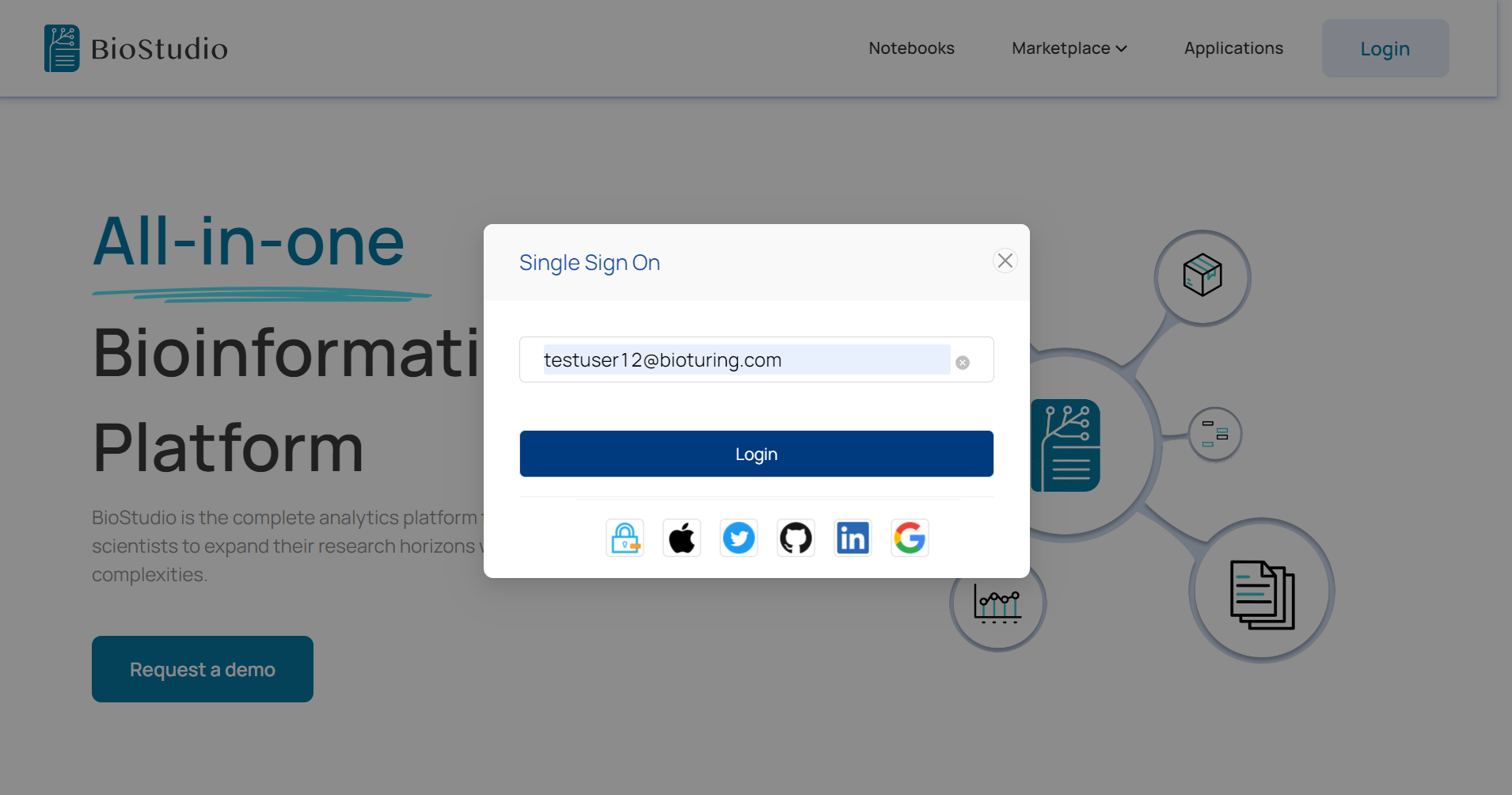
🔶 Login Succeed.
🔶 Before access WORKSPACE. It is necessary to access dashboard and create machine and well as create volume to be mounted.
🔗 Machine Creation and volume addition
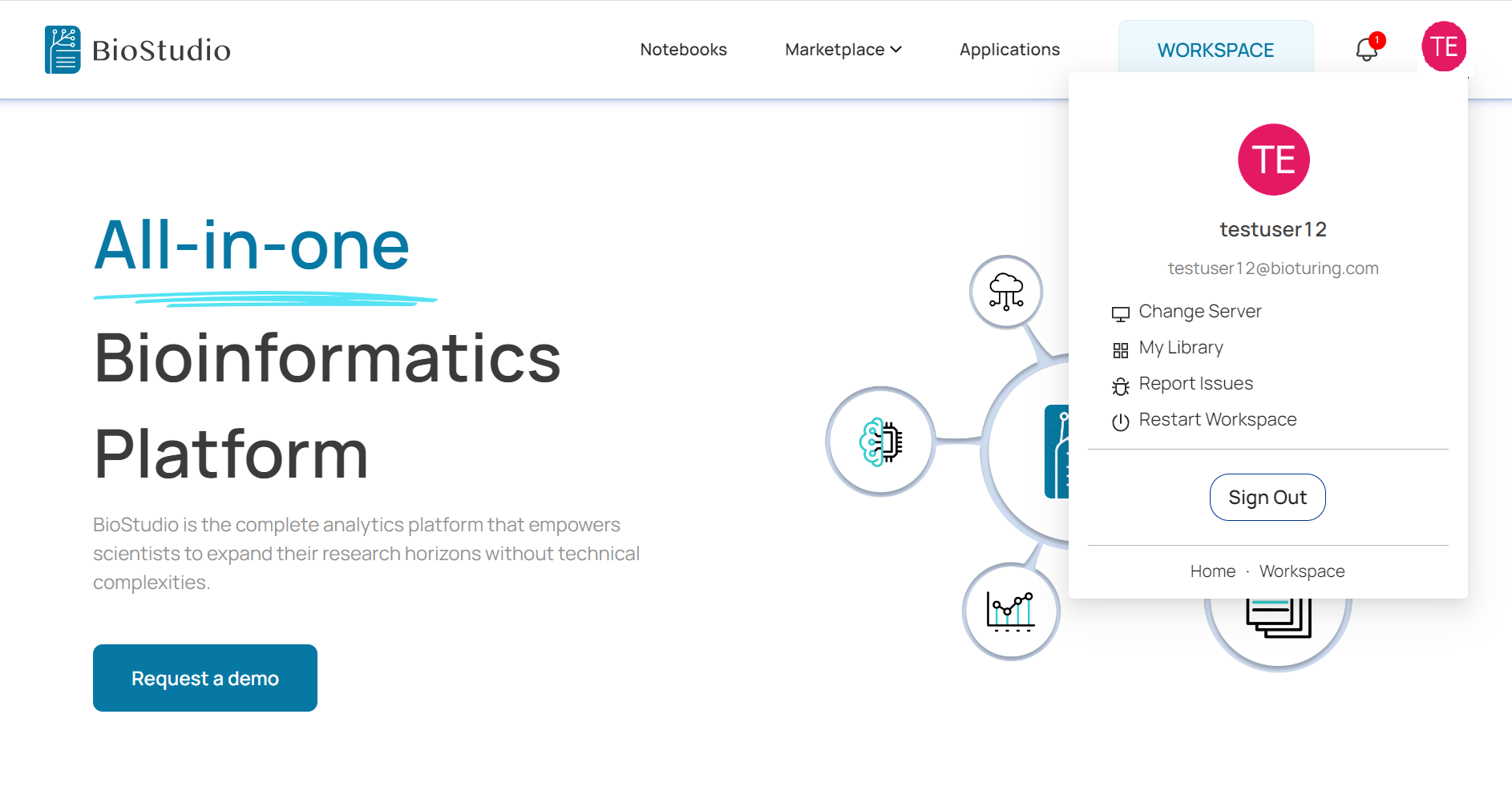
🔶 Access BioStudio services.
SSO set up with PingID ¶
🔶 Add an application to PingID
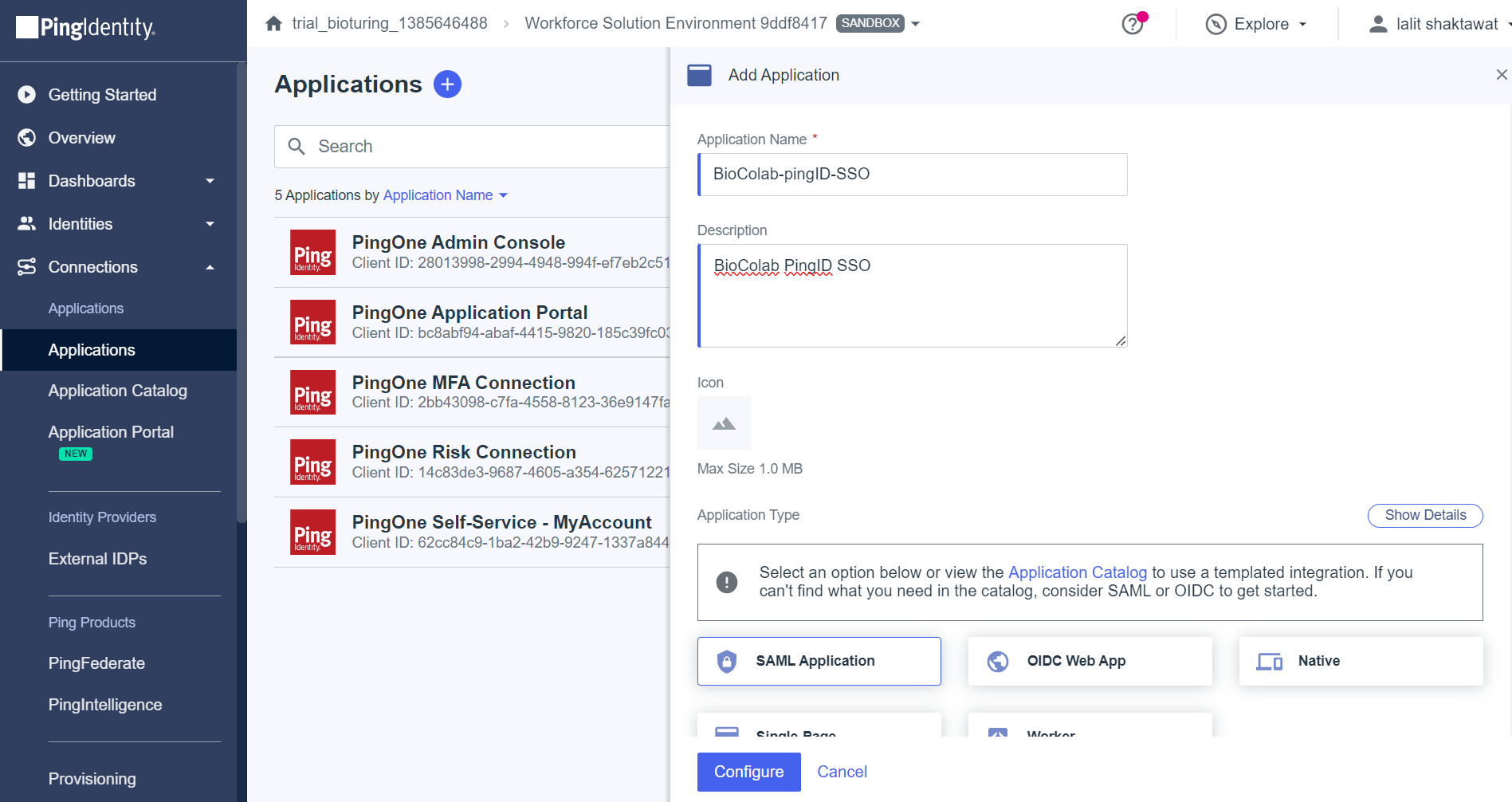
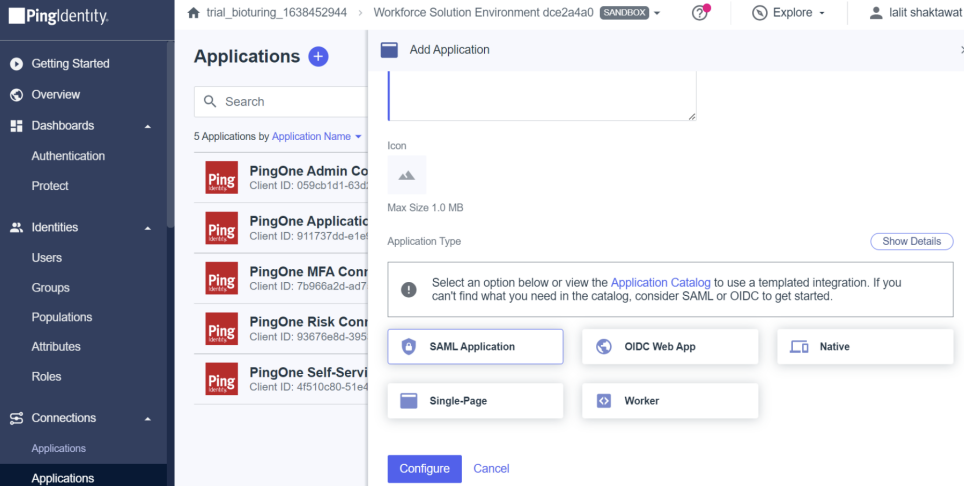
🔶 Select SAML Application method and click on Configure
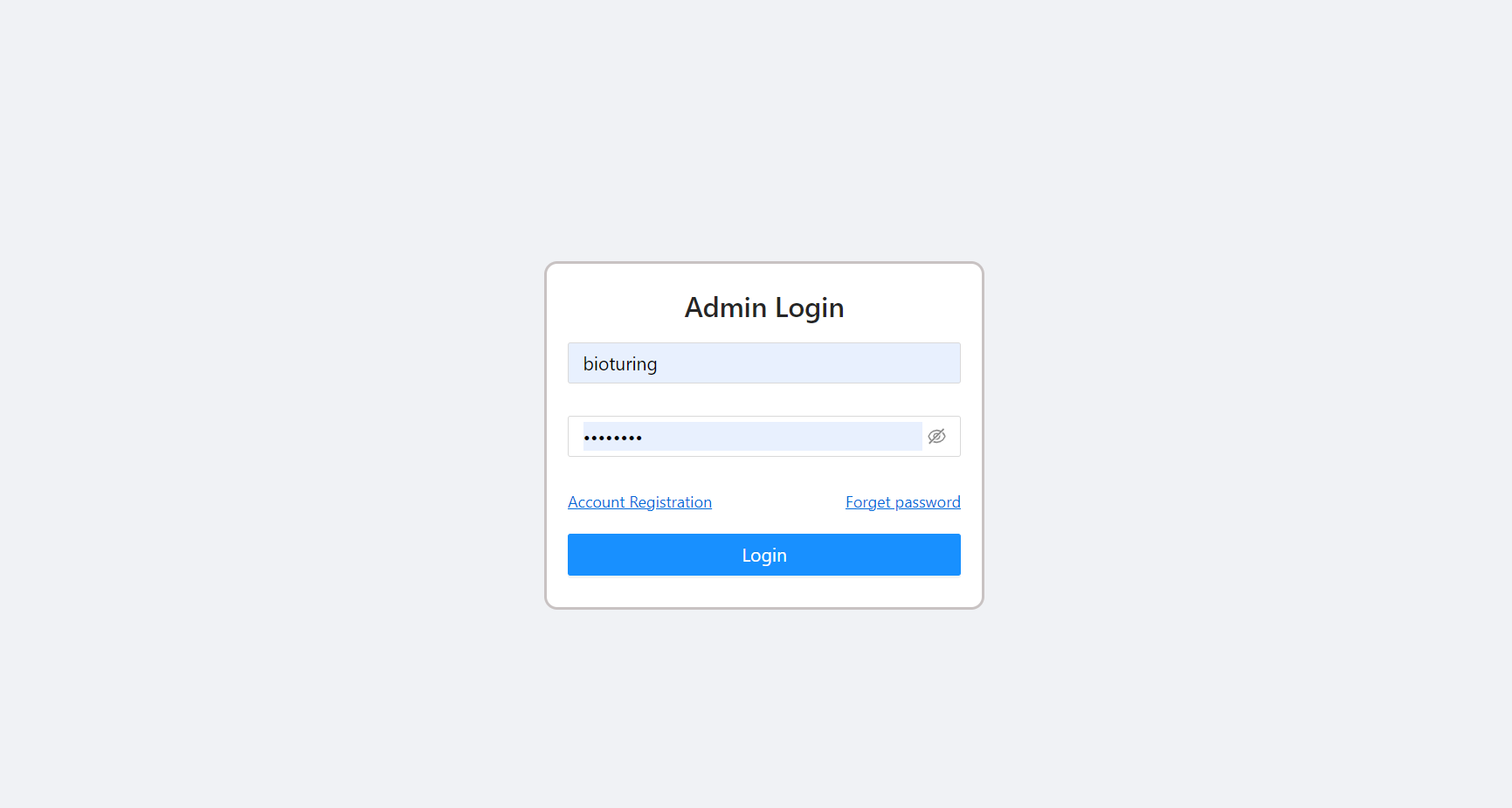
🔶 Login to BioColab Admin Dashboard
# https://<Domain name>/dashboard
It will prompt for Admin credentials. Please provide Admin credentials to login.
🔶 Select SSO setting from left side menu and click on + Add New SSO. If you are going to
set up SSO at the same time.
🔔 Note: If we are going to provide a Callback URL to another team to configure with IDP. Then we must provide dummy values to the mandatory field and click on submit button to save. Later we can update.
🔶 Copy Callback HTTPS URL that needs to be configured with IDP.
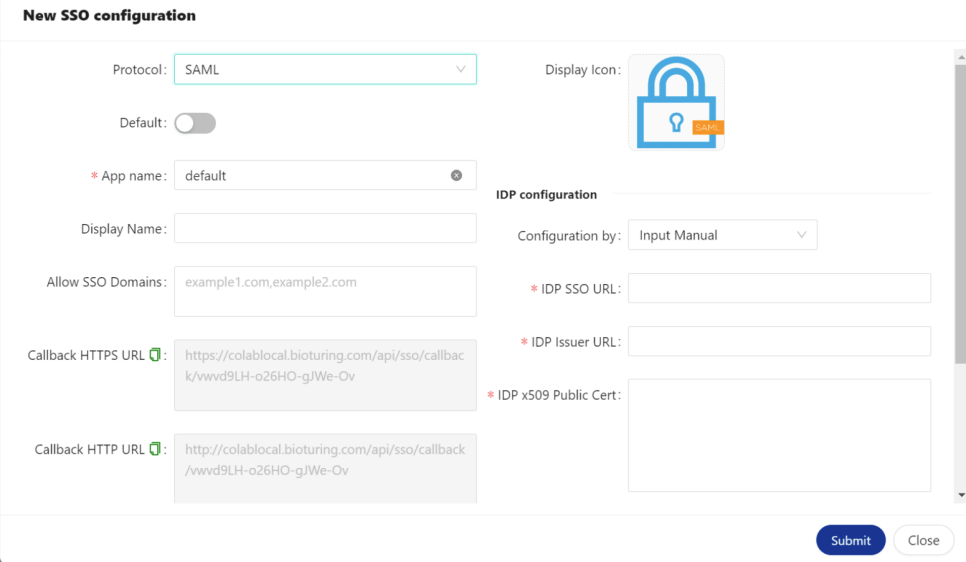
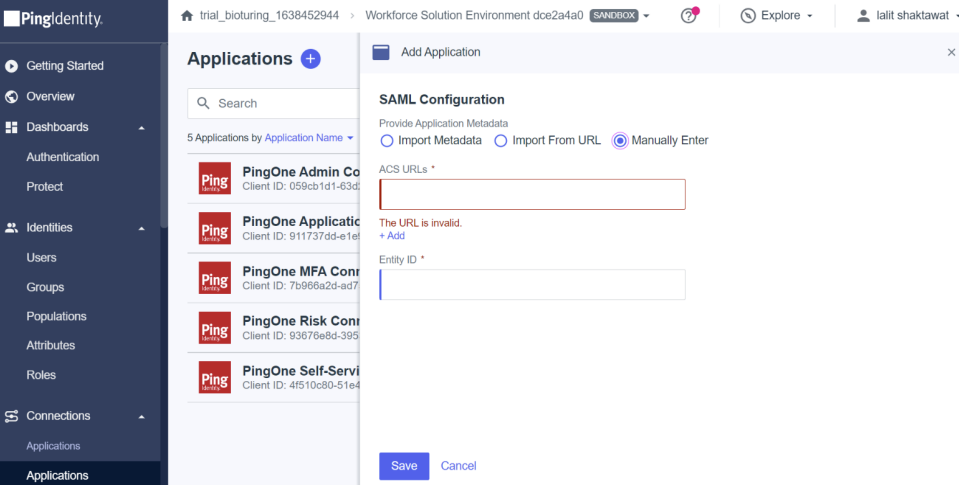
🔶 Select Manually Enter and fill callback URL to ACS URLs and Entity ID would be the last value of that URL.
🔶 Fill the value based on over instruction.
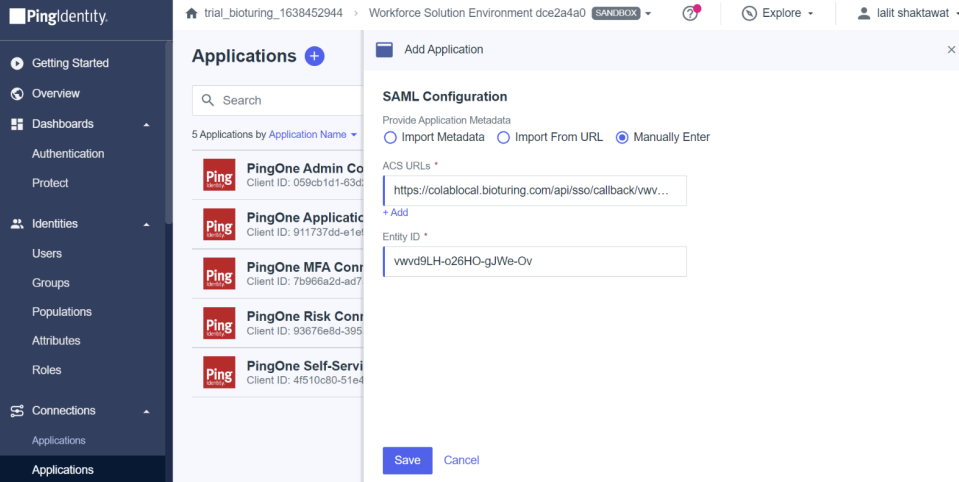
🔶 Click on Save button.
- Now update attribute mapping.
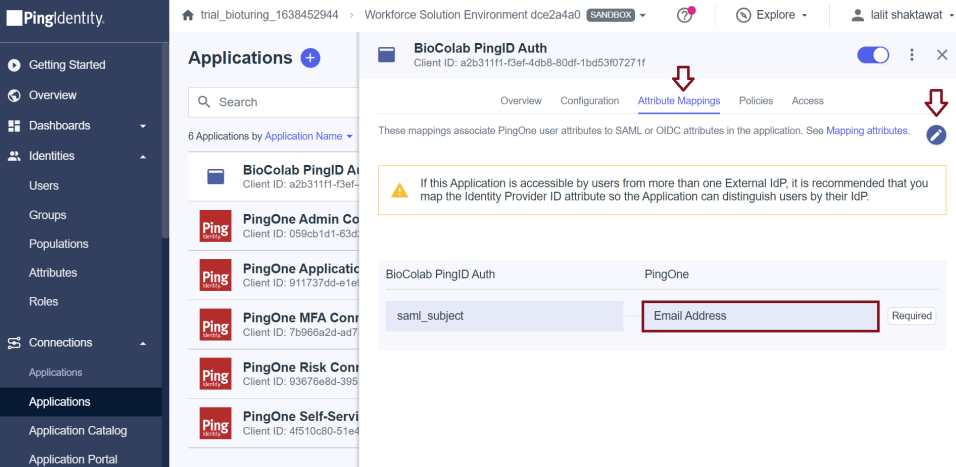
🔶 Select Configuration Tab and get all required values.
1️⃣ Issuer ID = IDP Issuer URL
2️⃣ Initiate Single Sign-On URL = IDP SSO URL
3️⃣ Download singing certificate = IDP x509 Public Cert
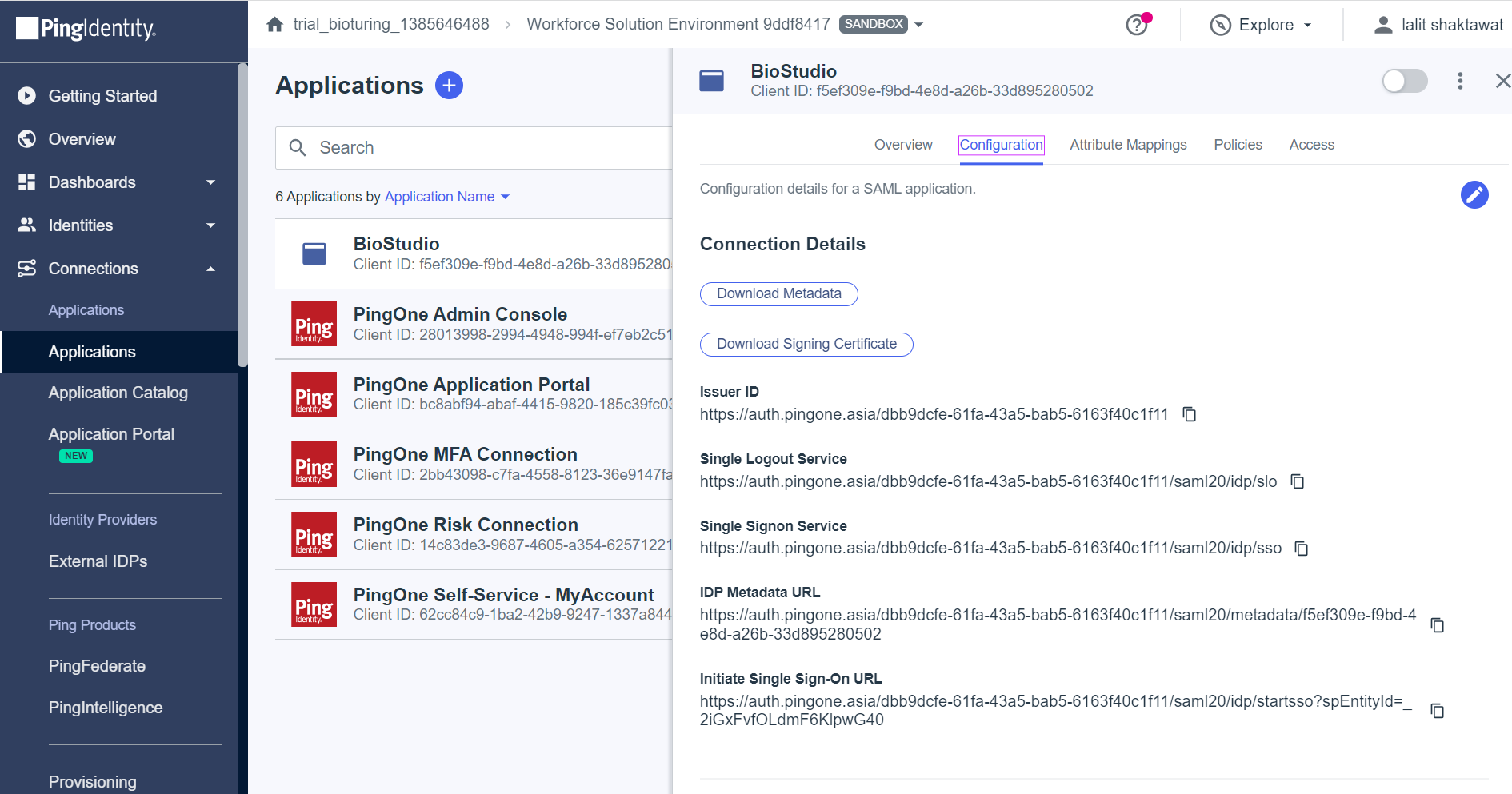
🔶 login to Biocolab dashboard admin and fill those values to required fields by updating SSO settings.
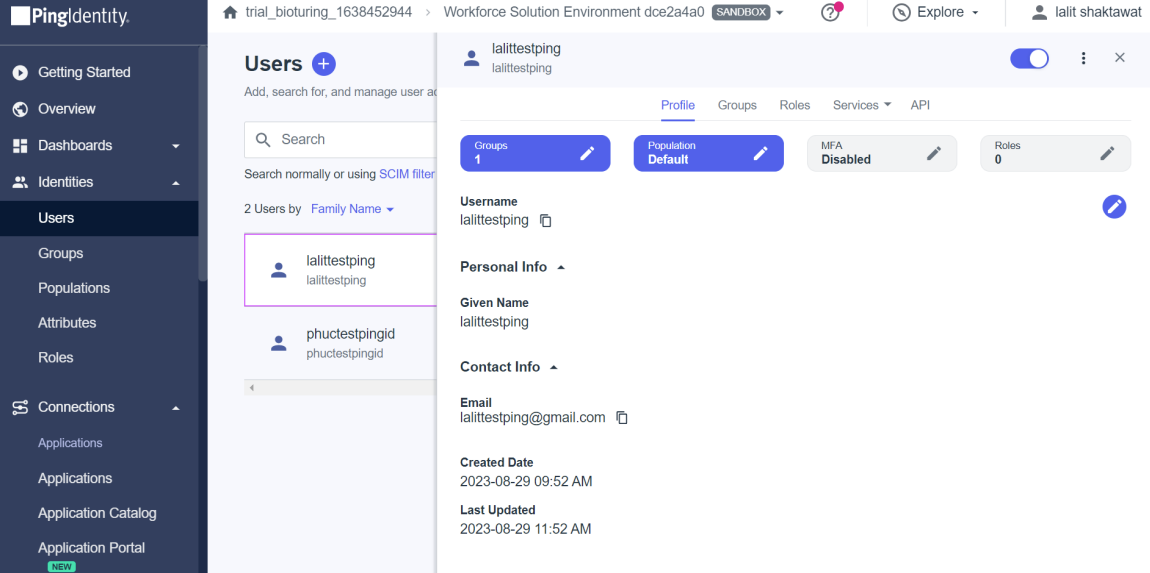
🔶 I created two users but on the existing set up you might have a user’s list already on the portal with a specified group to assign applications.
- On the Identity. I configured two users below.
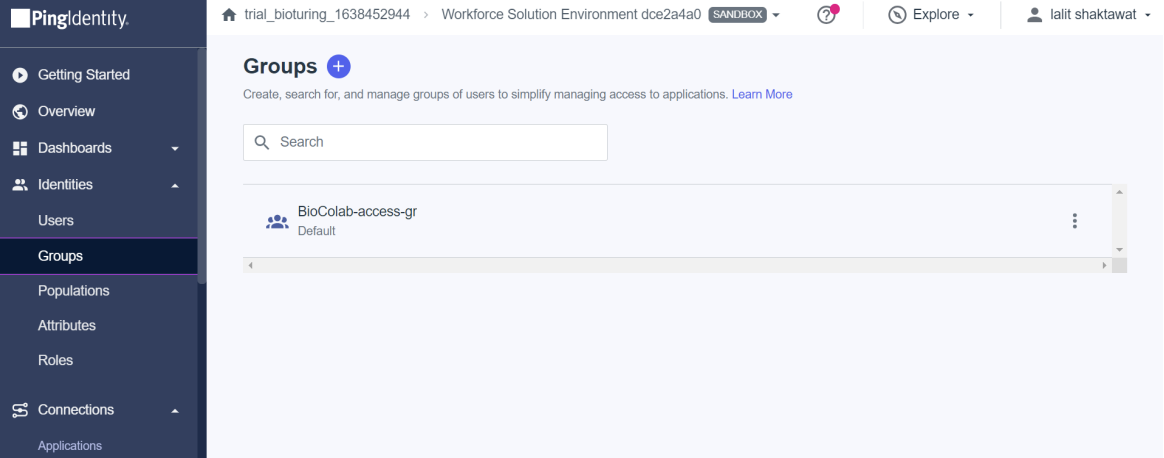
🔶 Now I created one Group and assigned those user’s to that Group
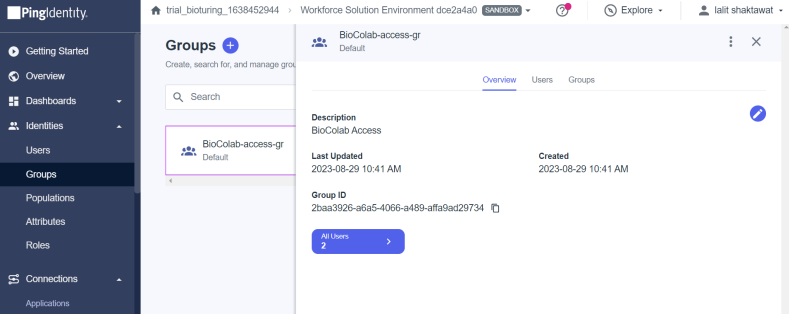
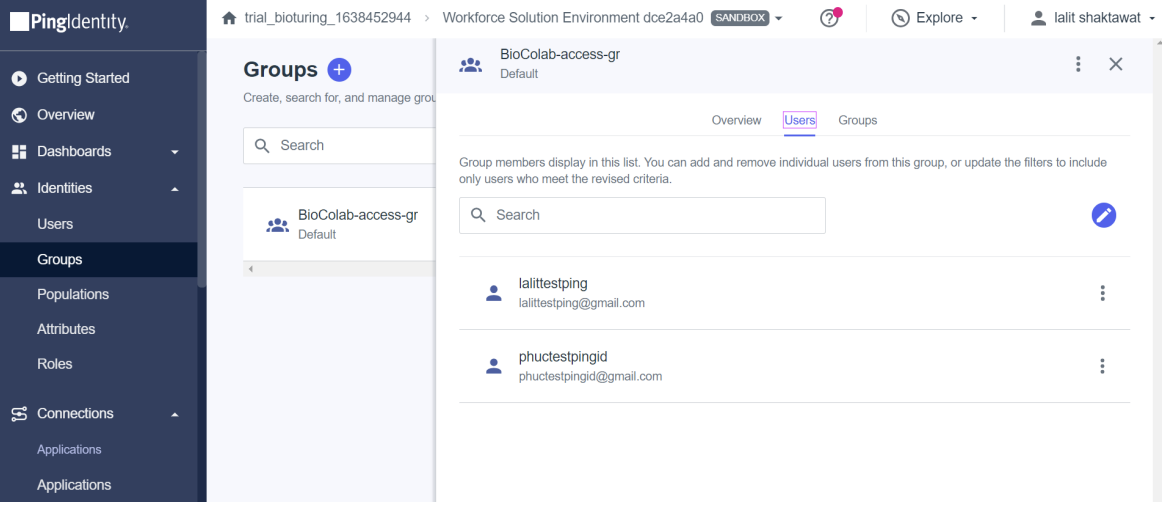
🔶 There are many ways to assign users to the group. Like User → Group → Edit – assign. Assign a group to the application.
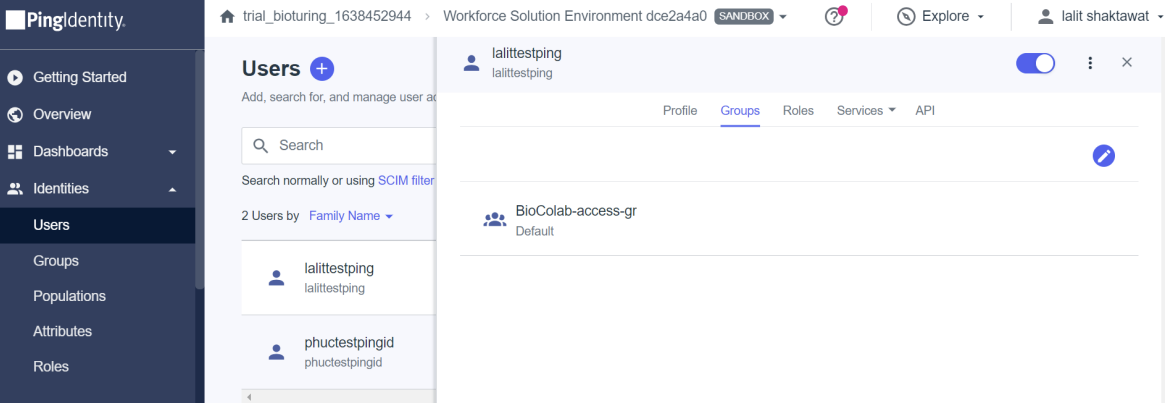
🔶 Now all set and time to test SSO login with those users.
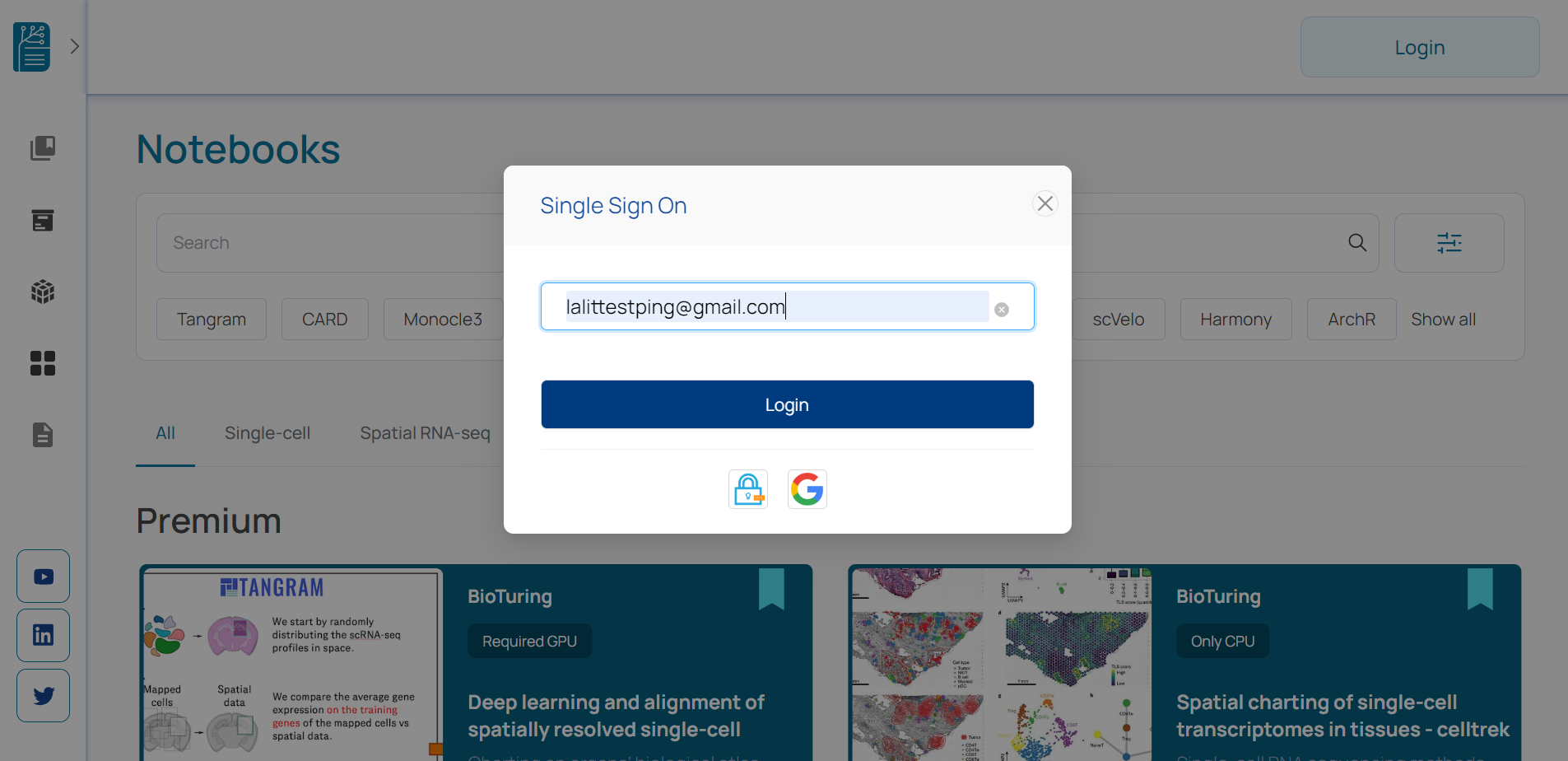
🔶 Provide PingID username and password, which we configured with Ping ID.
🔶 User is able to login successfully.
SSO set up with Azure AD ( SAML ) ¶
# Login to the dashboard admin
# https://<domain name>/dashboard
# provide admin credential, if already registered.
# If admin not yet registered, Kindly follow the instruction and registered first.
# NOTE: Make sure root password handy that you passed during BioStudio installation.
-
Dashboard login.
-
It will prompt for Admin credentials. Please provide Admin credentials to login.

🔶 Select SSO setting from left side menu and click on + Add New SSO. If you are going to
set up SSO at the same time.
🔔 Note: If we are going to provide a callback URL to another team to configure with IDP, Then we must provide dummy values to the mandatory field and click on the submit button to save. Later, we can update.

🔶 Copy Callback HTTPS URL that needs to be configured with IDP.
🔔 I am following to save the existing callback URL. Later, we will configure this.
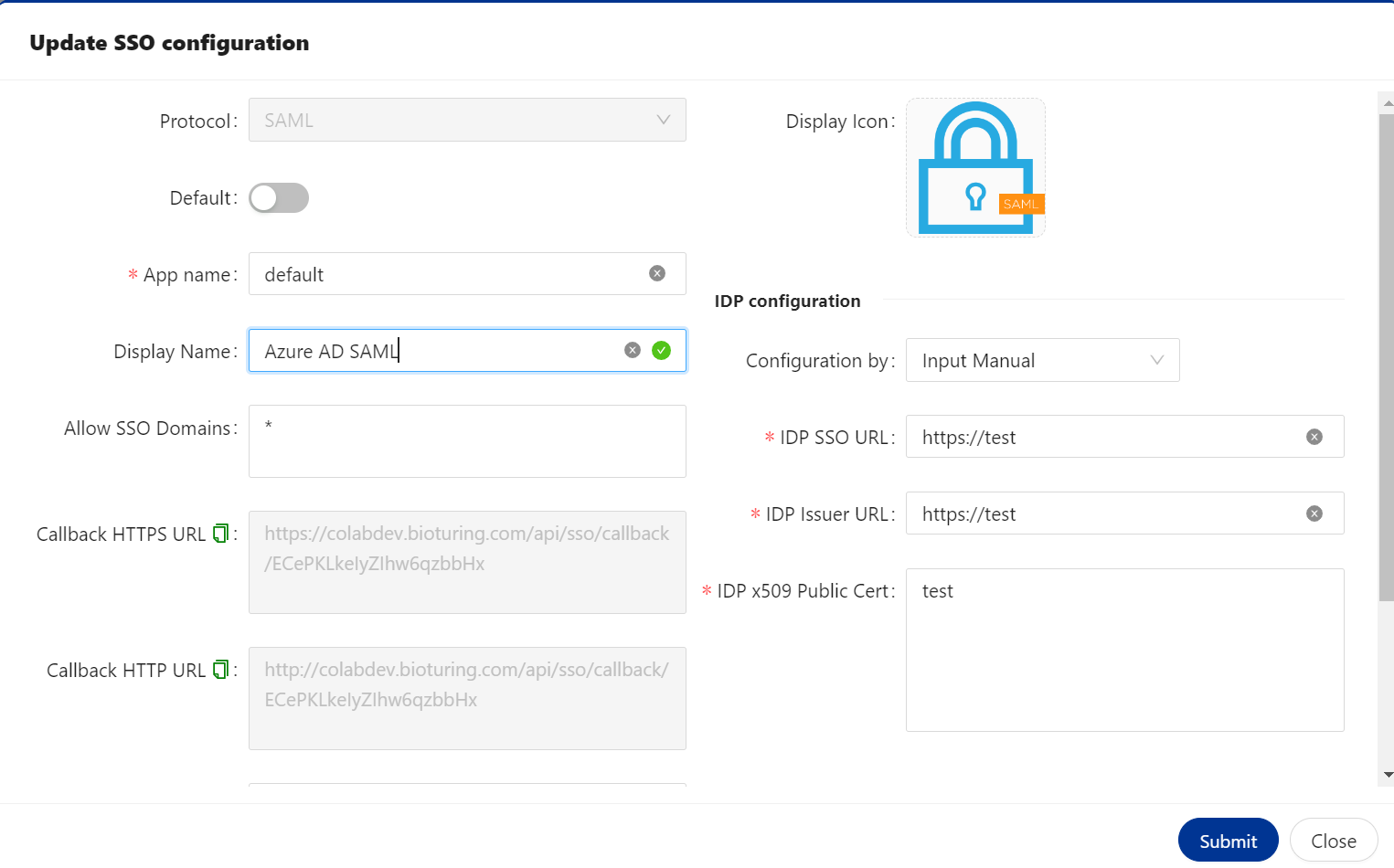
🔶 Login to Azure Cloud.
🔶 Open Microsoft Entra ID ( Azure Active Directory )
🔶 Click on Enterprise applications.
🔶 Click + New application.
🔶 Click + Create your own application.
🔶 Input your application name. Click on Create
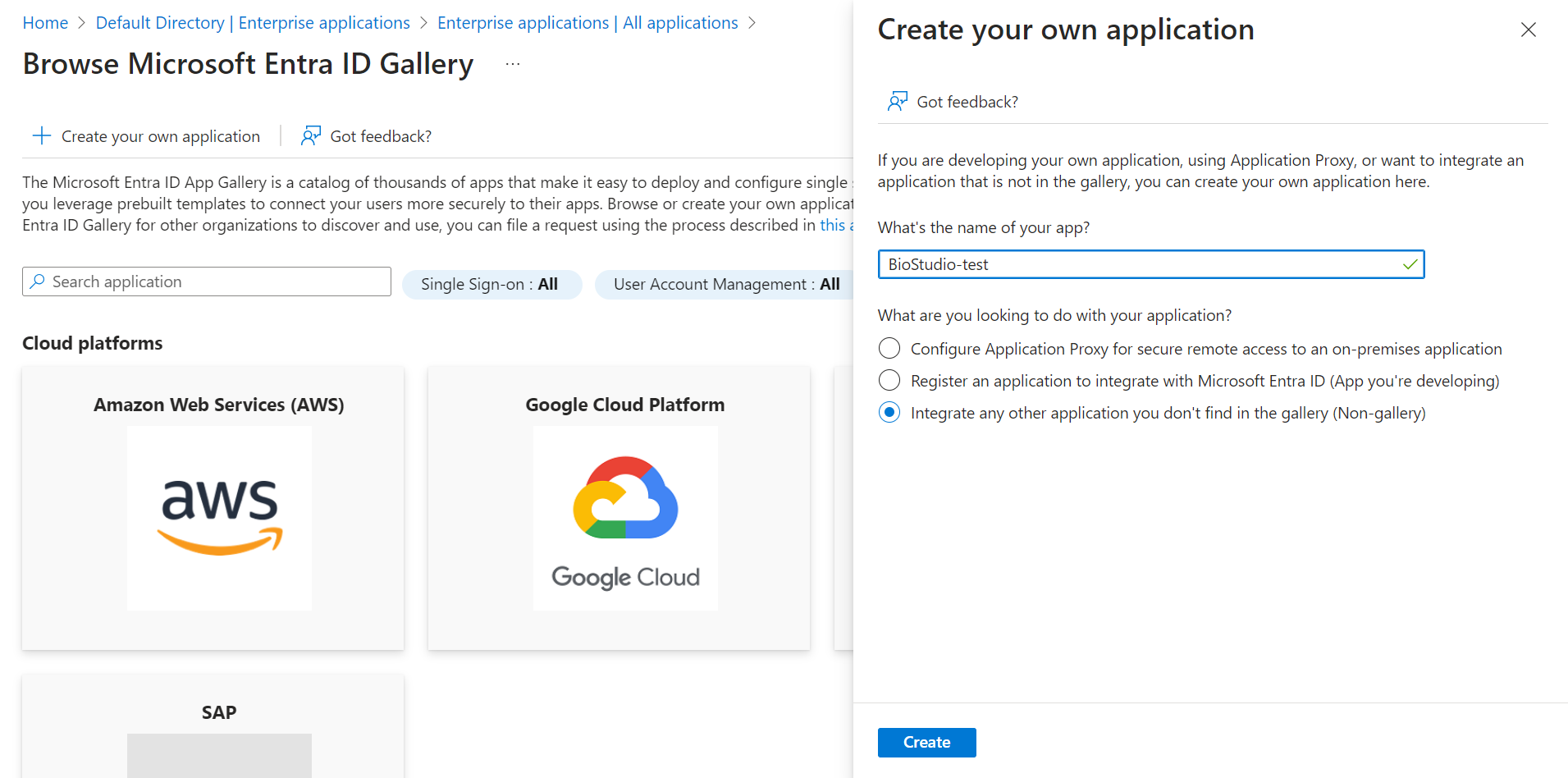
🔶 Application has been added.
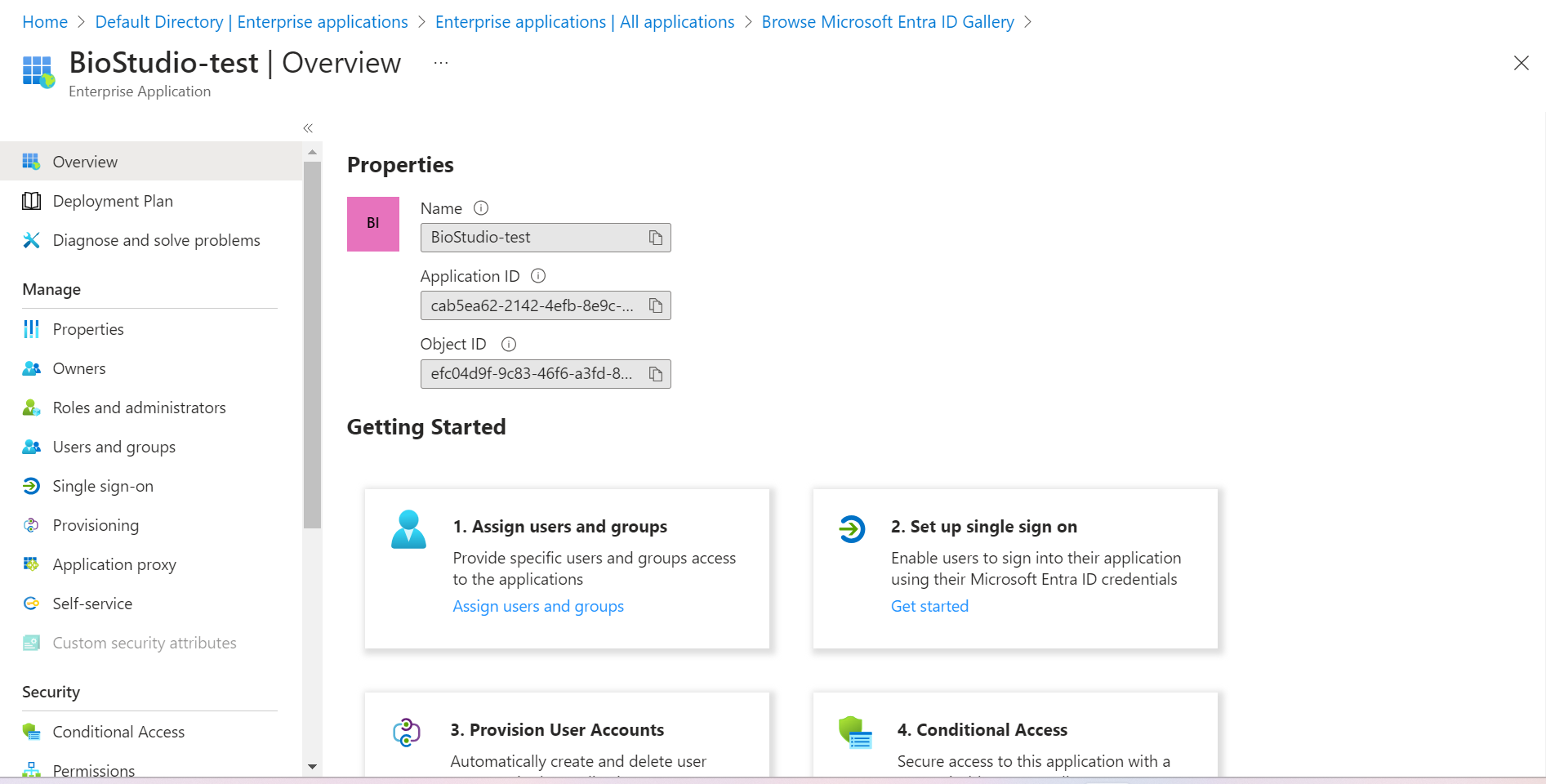
🔶 Click on Single sign-on.
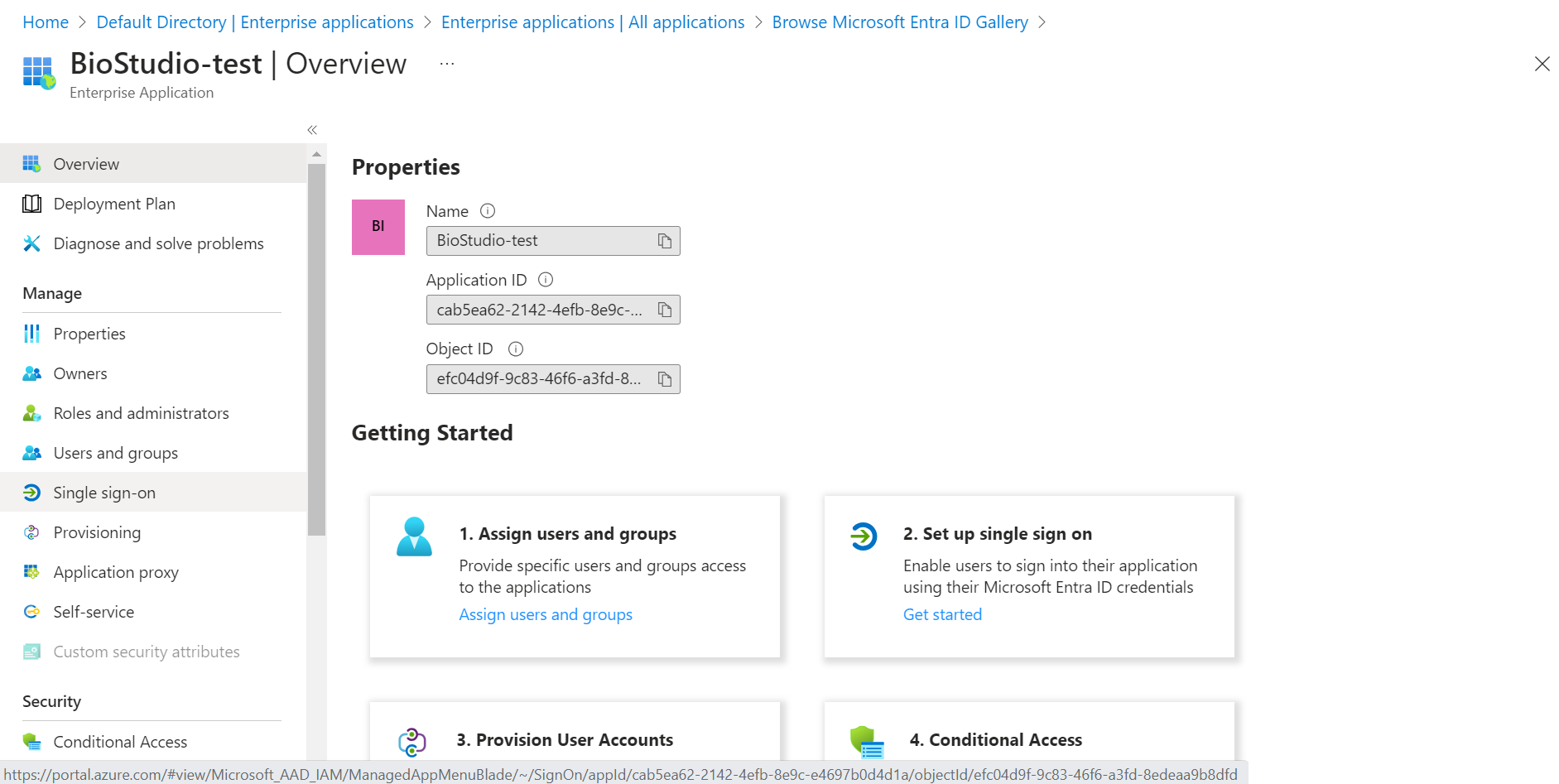
🔶 Click on SAML.
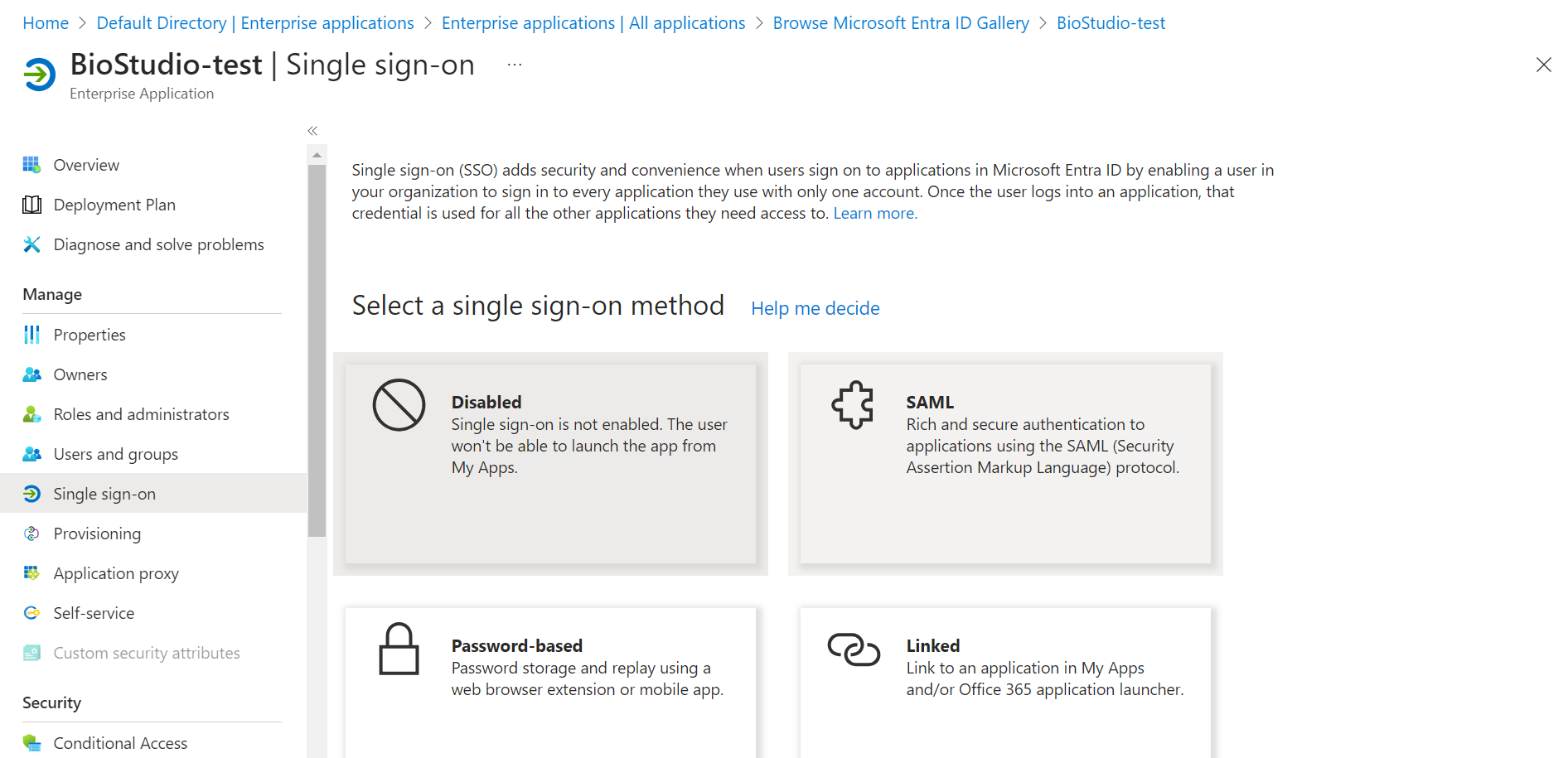
🔶 Click on Edit – Basic SAML Configuration
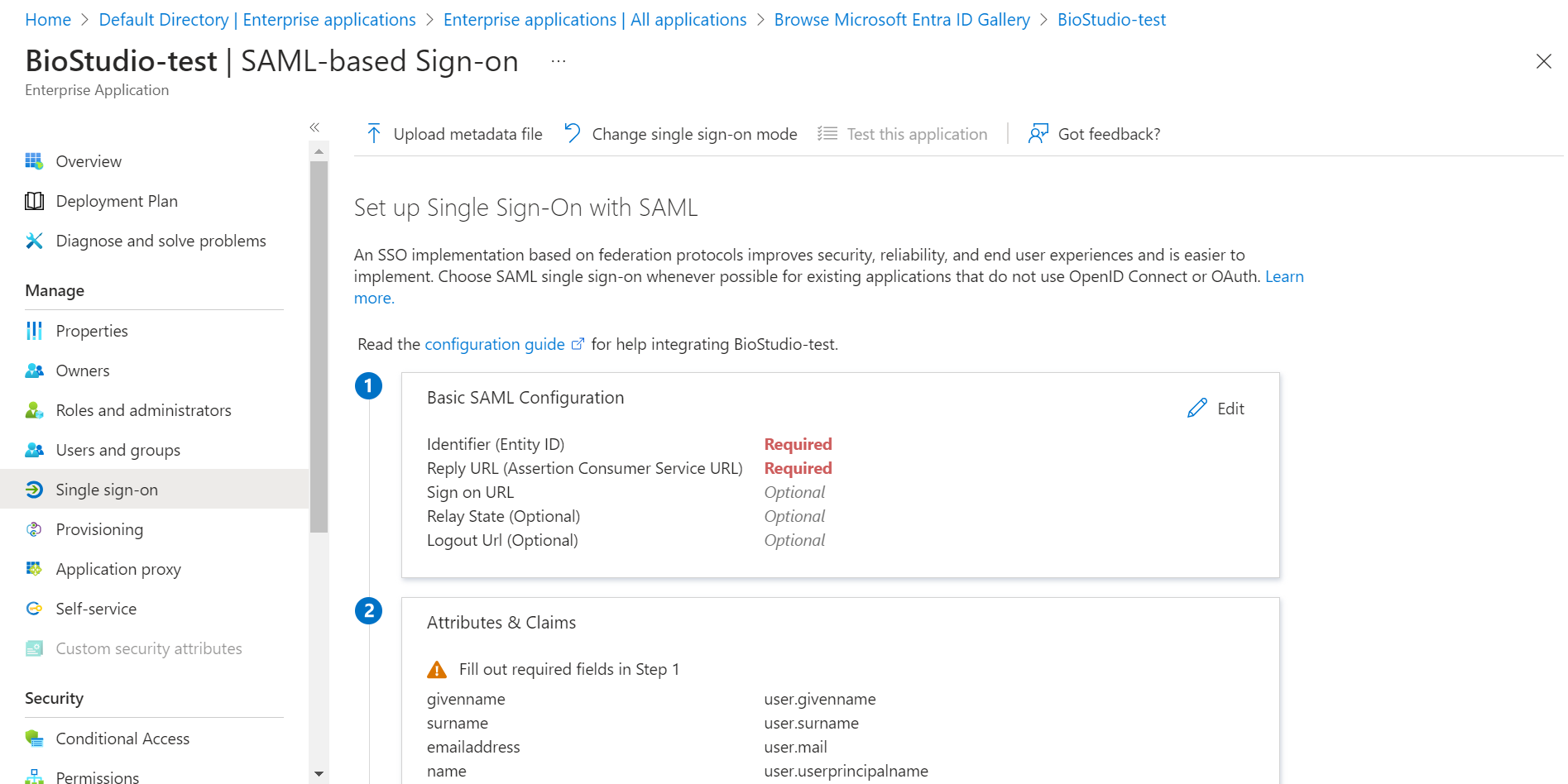
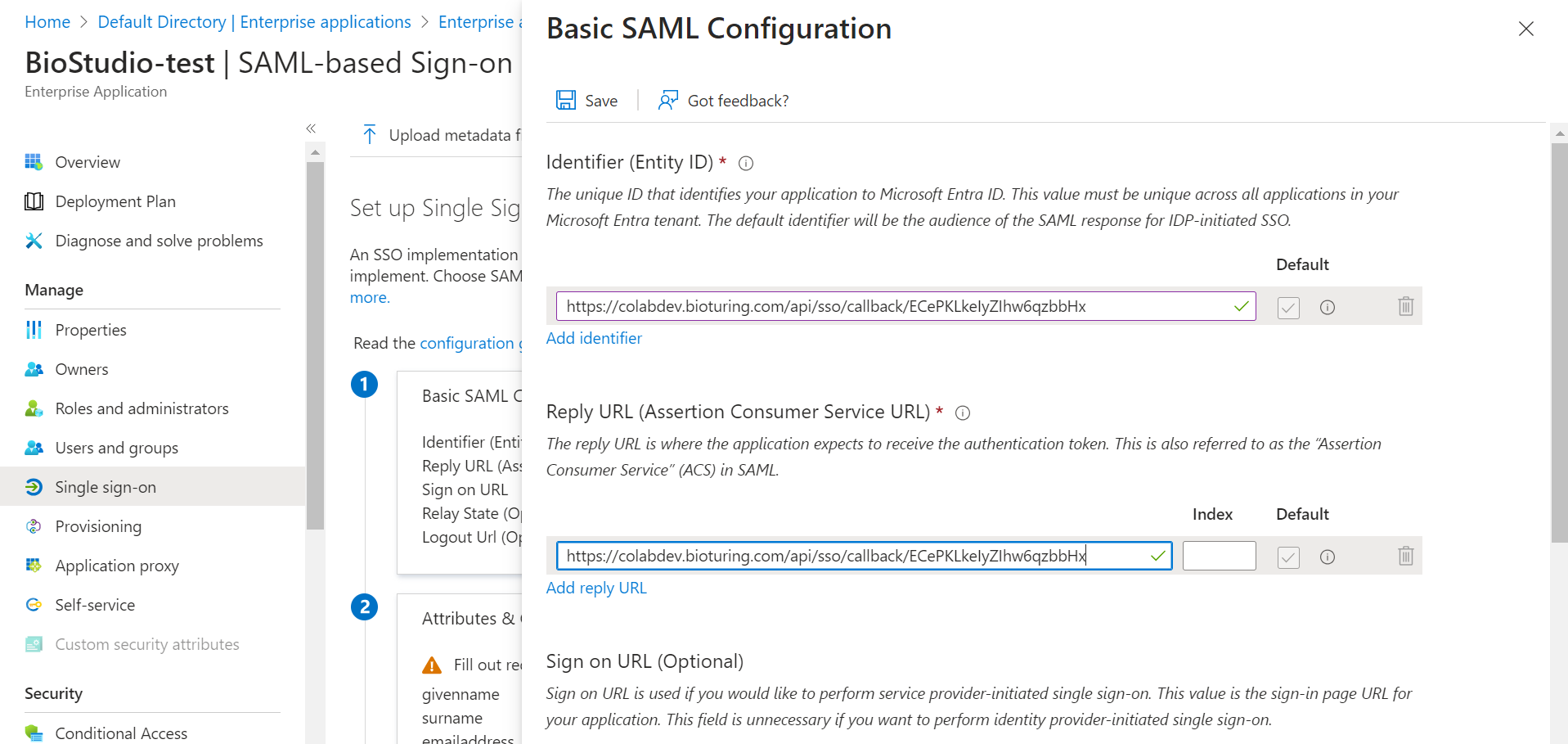
- Fill required field.
1️⃣ Identifier (Entity ID) : Callback URL
2️⃣ Reply URL (Assertion Consumer Service URL) : Callback URL
🔶 Login to BioStudio dashboard and click on three dot to select Update SSO setting
- Copy callback URL and fill on required fields.
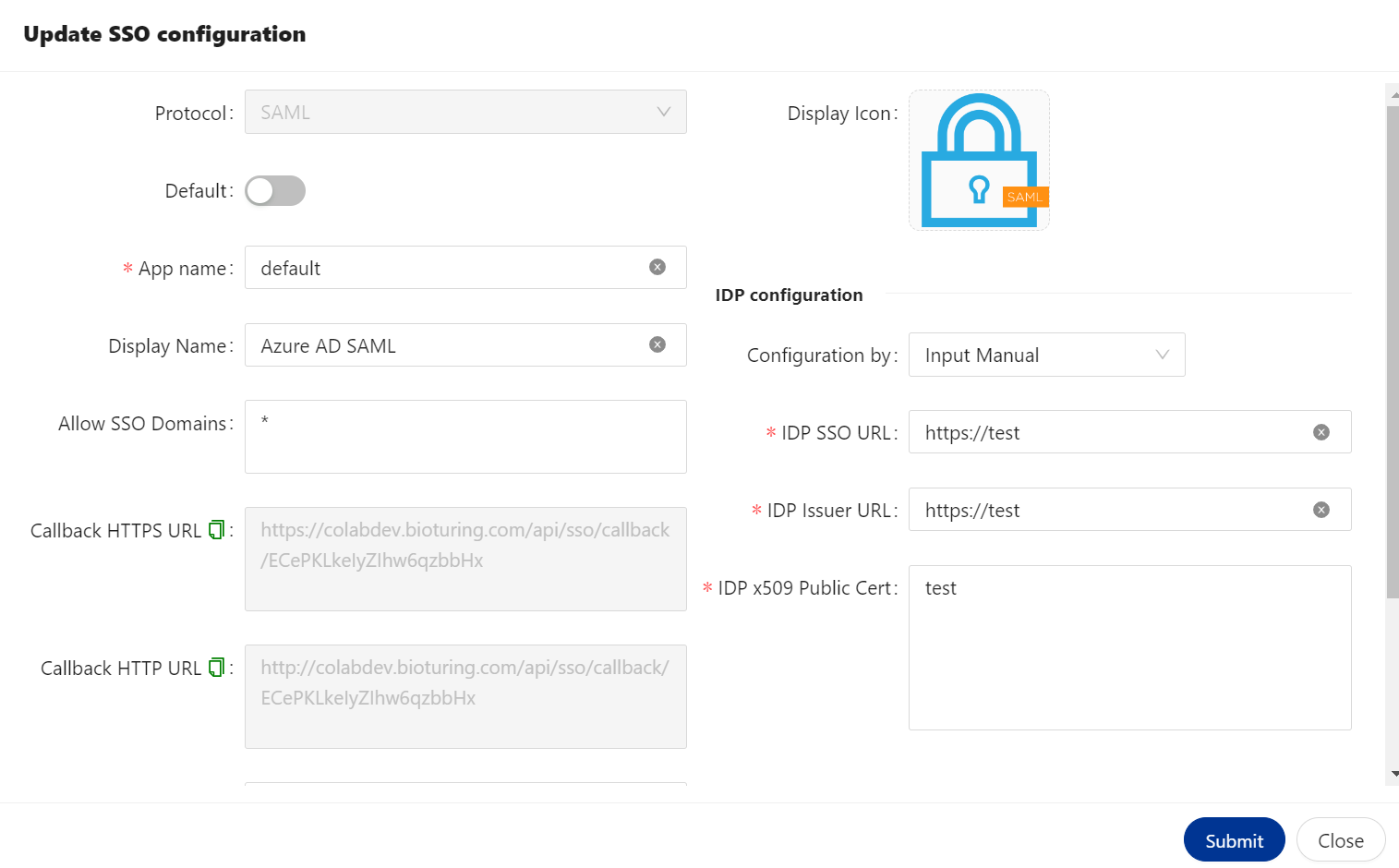
🔶 Click on Save button.
- Download Certificate (Base64)
- Copy Login URL
- Copy Microsoft Entra ID Identifier
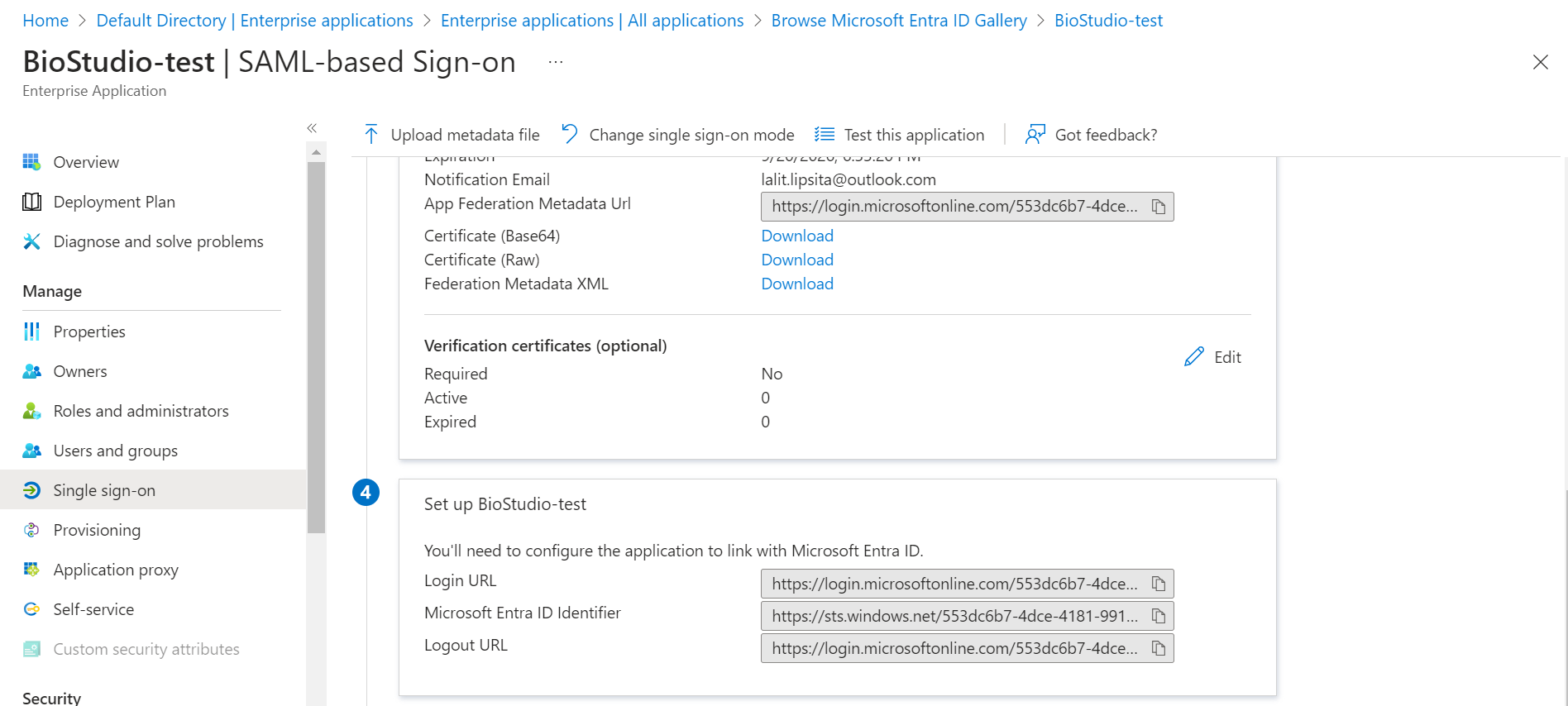
✏️ Certificate (Base64) = IDP x509 Public Cert
✏️ Login URL = IDP SSO URL
✏️ Microsoft Entra ID Identifier = IDP Issuer URL
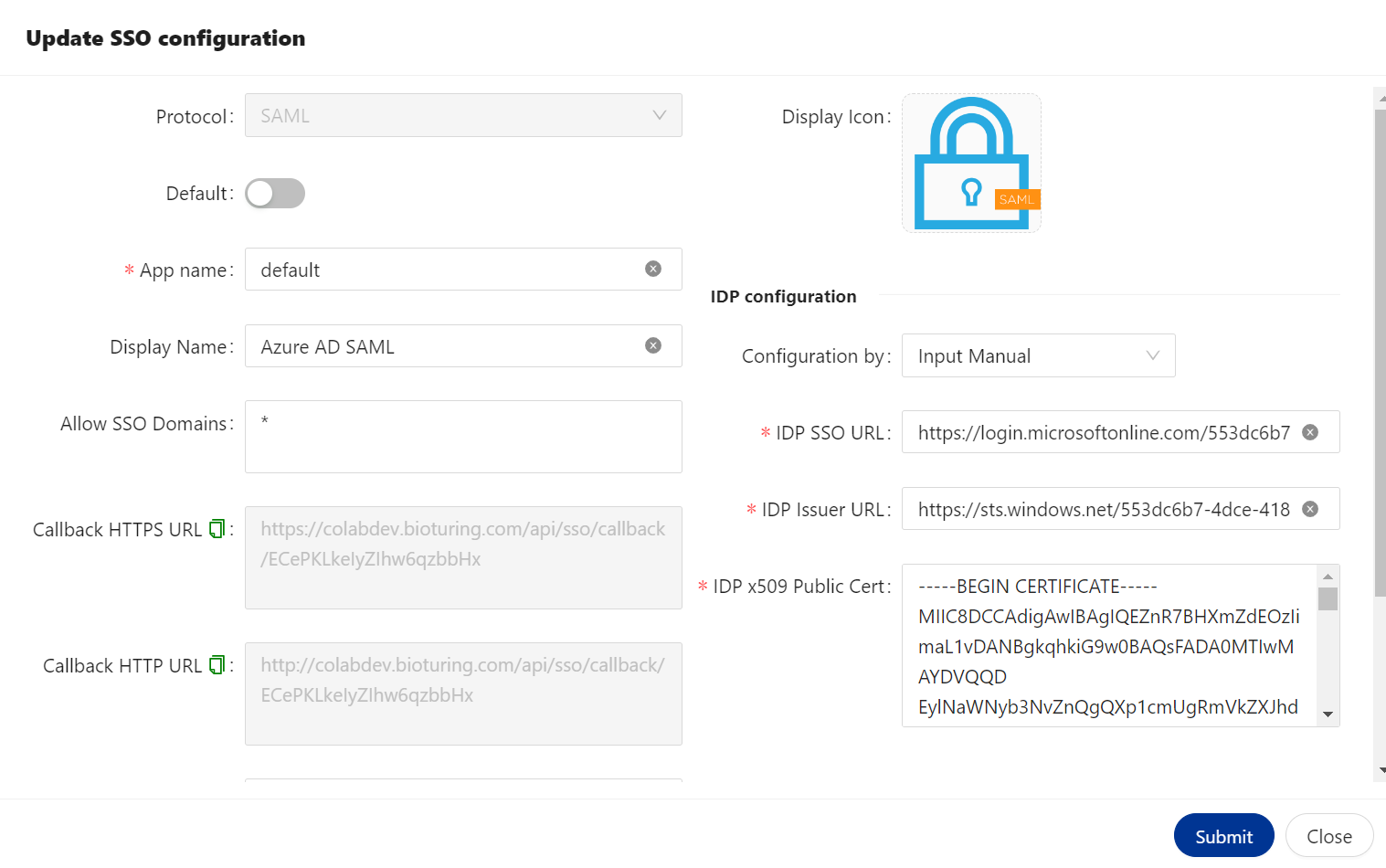
- Fill appropriate values to BioStudio SSO configuration.
🔶 Login to BioStudio dashboard and click on three dot to select Update SSO setting

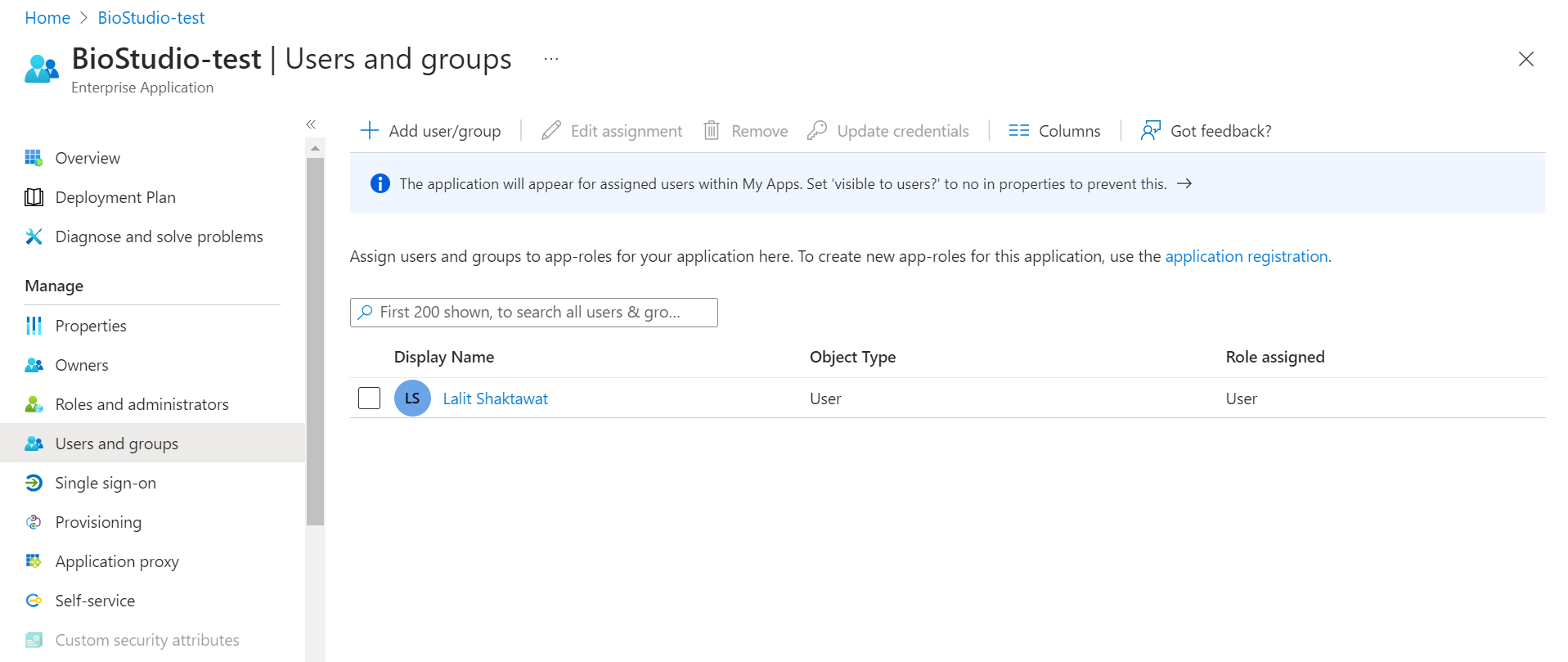
🔶 Create user and assign application to the user
🔶 Click on Users and groups
🔶 Click on + Add user/group
🔶 Click on Add user and click on Select
🔶 Click on Assign
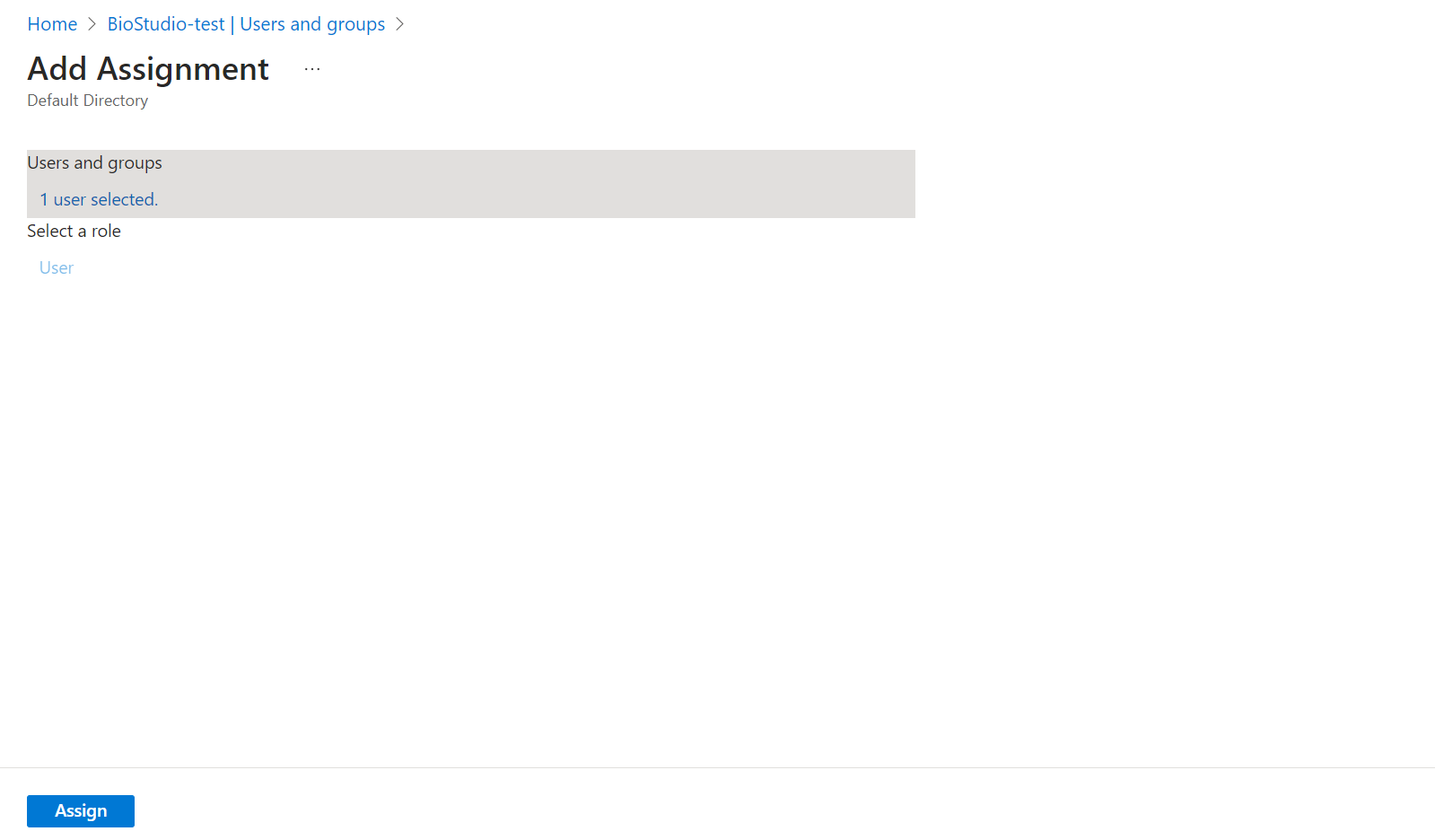
🔶 User has been Assign to this application.
🔶 Login succeed.
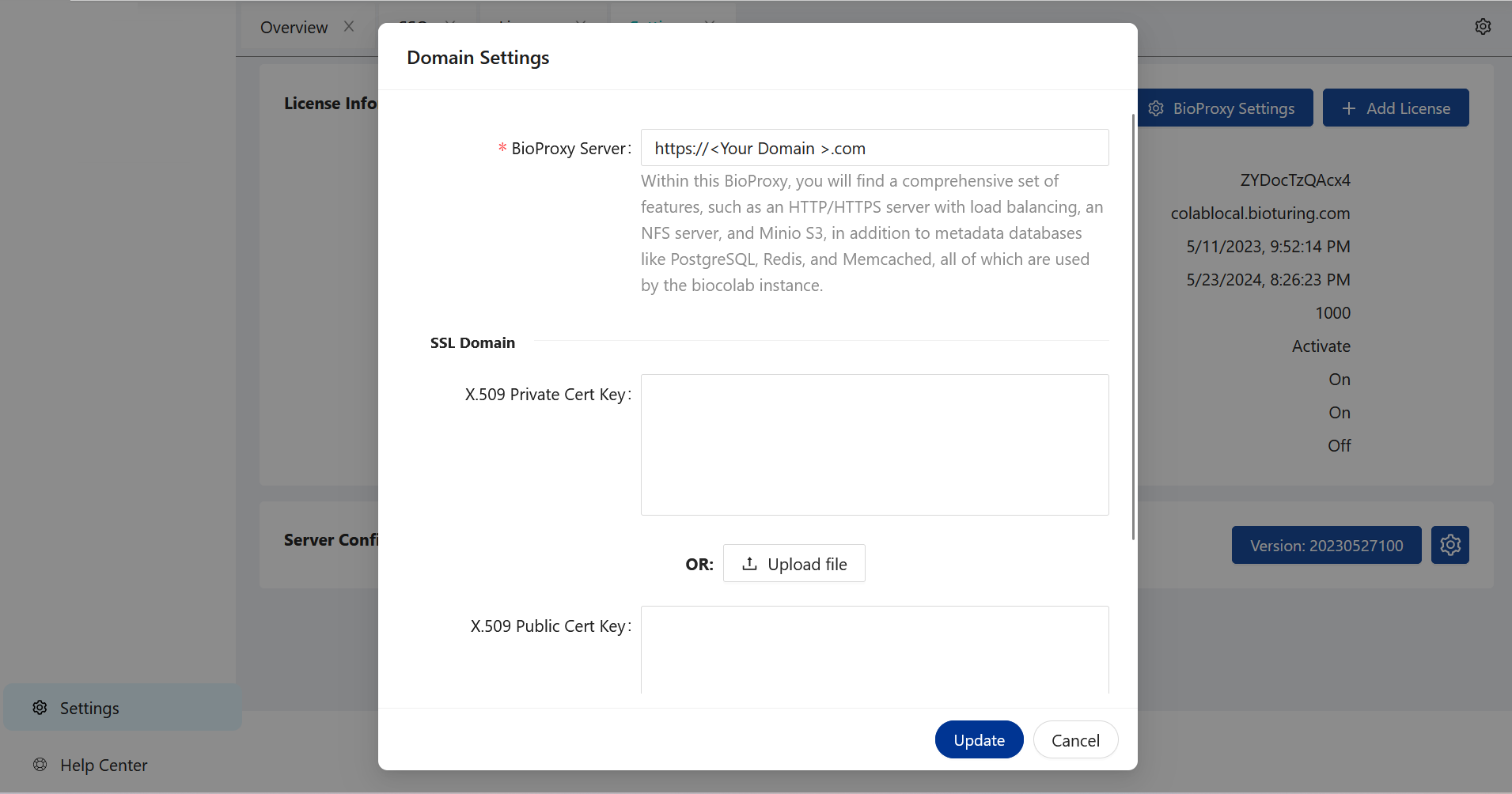
SSL setup. ¶
SSL Setup : Kindly get X.509 Provate and Public key.
I] Upload SSL file using web-ui.
# Just need tu put right value on write box and update.
# Or upload files as required.
# System will auto detect those updates.
II] Login to server and follow below steps.
# Copy tls.crt and tls.key file to /biocolab/configs.
# It was auto detected by our software and update the certificates.
🔶 SSL using WebUI
🔶 SSL direct upload on server location.

Kubernetes Setup for BioStudio ¶
helm search repo bioturing
NAME CHART VERSION APP VERSION DESCRIPTION
bioturing/biocolab 2.0.52 2.0.52 BioColab offers curated, ready-to-run notebooks...
bioturing/ecosystem 1.0.34 1.0.34 BioTuring System is a GPU-accelerated single-ce...
Helm chart version : 1.0.70
# Before installing the BioTuring System on Linux/K8S, some pre-installation steps are required:
# System: K8s
# A token obtained from BioTuring for BioStudio.
# BioStudio supporting GPU and CPU type of instance / Machine.
# Kindly select GPU, If you wish to use Prebuilt GPU based notebook or wish to generate GPU based note book.
Prepare GPU toolkit for K8S ¶
Patch container engines (Docker, Containerd)
Install NVidia container toolkit on each node following the guide:
🔗 nvidia toolkit install guide
🔶 Check container engines (Docker, Containerd)
# For microk8s :
microk8s kubectl describe no | grep Runtime
# For vanilla :
kubectl describe no | grep Runtime
▶️ If container engine is Containerd, add these lines to : /etc/containerd/config.toml
privileged_without_host_devices = false
base_runtime_spec = ""
[plugins."io.containerd.grpc.v1.cri".containerd.runtimes.runc.options]
SystemdCgroup = true
[plugins."io.containerd.grpc.v1.cri".containerd.runtimes.nvidia]
privileged_without_host_devices = false
runtime_engine = ""
runtime_root = ""
runtime_type = "io.containerd.runc.v1"
[plugins."io.containerd.grpc.v1.cri".containerd.runtimes.nvidia.options]
BinaryName = "/usr/bin/nvidia-container-runtime"
SystemdCgroup = true
[plugins."io.containerd.grpc.v1.cri".cni]
bin_dir = "/opt/cni/bin"
conf_dir = "/etc/cni/net.d"
▶️ After that, restart containerd
sudo systemctl restart containerd
sudo nvidia-container-cli --load-kmods info
If container engine is Docker, add these lines to: /etc/docker/daemon.json
{
"default-runtime": "nvidia",
"runtimes": {
"nvidia": {
"path": "nvidia-container-runtime",
"runtimeArgs": []
}
}
}
▶️ After that, restart docker
sudo systemctl restart docker
sudo nvidia-container-cli --load-kmods info
Install BioTuring ecosystem on K8S ¶
We support all k8s engines: GKE (Google Kubernetes Engine), EKS (Amazon Elastic Kubernetes Service), AKS (Azure Kubernetes Service), MicroK8s, and vanilla K8S …etc.
Ensure that helm (version 3) is installed.
▶️ First, check the Helm version.
# Example :
microk8s enable helm3
microk8s helm3 version
Helm command tips:
# Add repo charts
helm repo add bioturing https://bioturing.github.io/charts/apps/
helm repo update
helm search repo bioturing
# Login BioTuring registry
helm registry login -u admin registry.bioturing.com
# Show information of helm chart name
helm show all bioturing/<helm chart name> --version <helm chart version>
# Action into helm chart name
helm template bioturing bioturing/<helm chart name> --version <helm chart version>
helm install bioturing bioturing/<helm chart name> --version <helm chart version>
helm upgrade bioturing bioturing/<helm chart name> --version <helm chart version>
⭕ Add BioTuring Helm charts
# Example:
For Vanilla K8s:
helm repo add bioturing https://bioturing.github.io/charts/apps/
For Microk8s:
microk8s helm3 repo add bioturing https://bioturing.github.io/charts/apps/
Helm Chart ¶
| Key | Type | Default | Description |
|---|---|---|---|
| image.biocolab.repository | string | bioturing/biocolab | biocolab repository |
| image.biocolab.tag | int | 2.0.50 | biocolab image tag |
| image.bioproxy.repository | string | bioturing/biocolab | bioproxy repository |
| image.bioproxy.tag | int | 1.0.26 | bioproxy image tag |
| imagePullSecrets | object | {} | secrets |
| secret.data.allowips | string | "" | allow ip |
| secret.data.cbtoken | string | "" | Colab token |
| secret.data.domain | string | "" | domain name |
| secret.admin.username | string | admin | admin user name |
| secret.admin.password | string | passwdadmin12 | admin password |
| secret.postgresql.dbcolab | string | "" | postgres DB name |
| secret.postgresql.dbhub | string | "" | postgres hub DB name |
| secret.postgresql.username | string | "" | postgres user name |
| secret.postgresql.password | string | "" | postgres password |
| secret.server.aria2c_list_ips | object | {} | |
| secret.server.certificate | string | "" | Server certificate |
| secret.server.key | string | "" | Server Key |
| secret.server.collaborative_mode | bool | false | |
| secret.server.debug_mode | bool | false | |
| secret.server.enable_https | bool | false | |
| secret.server.memcached_list | string | "" | |
| secret.server.mqtt_list_ips | string | "" | |
| secret.server.redis_list | string | "" | |
| secret.server.redis_password | string | "" | |
| secret.server.tracing_mode | bool | false | |
| secret.server.trakfik_proxy_mode | bool | false | |
| secret.server.use_letsencrypt | bool | false | |
| secret.server.use_redis_cache | bool | false | |
| service.ports.biocolab.aria2c.port | int | 6800 | |
| service.ports.biocolab.http.port | int | 11123 | application port |
| service.ports.biocolab.jobqueue.port | int | 11300 | |
| service.ports.biocolab.mqtttcp.port | int | 1883 | |
| service.ports.biocolab.mqttweb.port | int | 9001 | |
| service.ports.biocolab.notebook.port | int | 18000 | |
| service.ports.bioproxy.http.port | int | 80 | http port |
| service.ports.bioproxy.https.port | int | 443 | https port |
| service.ports.bioproxy.memcached.port | int | 11211 | |
| service.ports.bioproxy.ntfsp1.port | int | 111 | |
| service.ports.bioproxy.ntfsp2.port | int | 2049 | |
| service.ports.bioproxy.ntfsp3.port | int | 32767 | |
| service.ports.bioproxy.ntfsp4.port | int | 32765 | |
| service.ports.bioproxy.postgresql.port | int | 5432 | |
| service.ports.bioproxy.redis.port | int | 6379 | redis port |
| persistence.dirs.app.size | String | 150Gi | application data |
| persistence.dirs.app.storageClass | string | "" | |
| persistence.dirs.metadata.size | String | 5Gi | metadata |
| persistence.dirs.metadata.storageClass | string | "" | |
| persistence.dirs.user.size | String | 500Gi | user data |
| persistence.dirs.user.storageClass | string | "" | |
| ingress.annotations | object | {} | |
| ingress.className | string | "" | |
| ingress.classNginxName | string | nginx | |
| ingress.enabled | bool | true | |
| ingress.tls | bool | true | |
| ingress.tls.enabled | bool | true | |
| ingress.useNginx | bool | false | |
| nodeSelector | object | {} | |
| podAnnotations | object | {} | |
| podSecurityContext | object | {} | |
| replicaCount | int | 1 | |
| resources | object | {} | |
| service.type | string | ClusterIP | |
| serviceAccount.annotations | object | {} | |
| serviceAccount.name | string | "" | |
| tolerations | object | {} | |
| host_ip | int | 0.0.0.0 | |
| affinity | object | {} | |
| autoscaling | object | {} | |
| autoscaling.enabled | bool | false | |
| autoscaling.maxReplicas | int | 100 | |
| autoscaling.minReplicas | int | 2 | |
| autoscaling.targetCPUUtilizationPercentage | int | 80 | |
| gpu.runtimeClassName | string | "" |
🔶 Select helm chart version.
⭕Kindly select latest version of Helm Chart. Support team will get in touch with you and share latest version tag.
# Click on install.

🔶 Select namespace and type name of Statefulset.
🔶 Click on Next
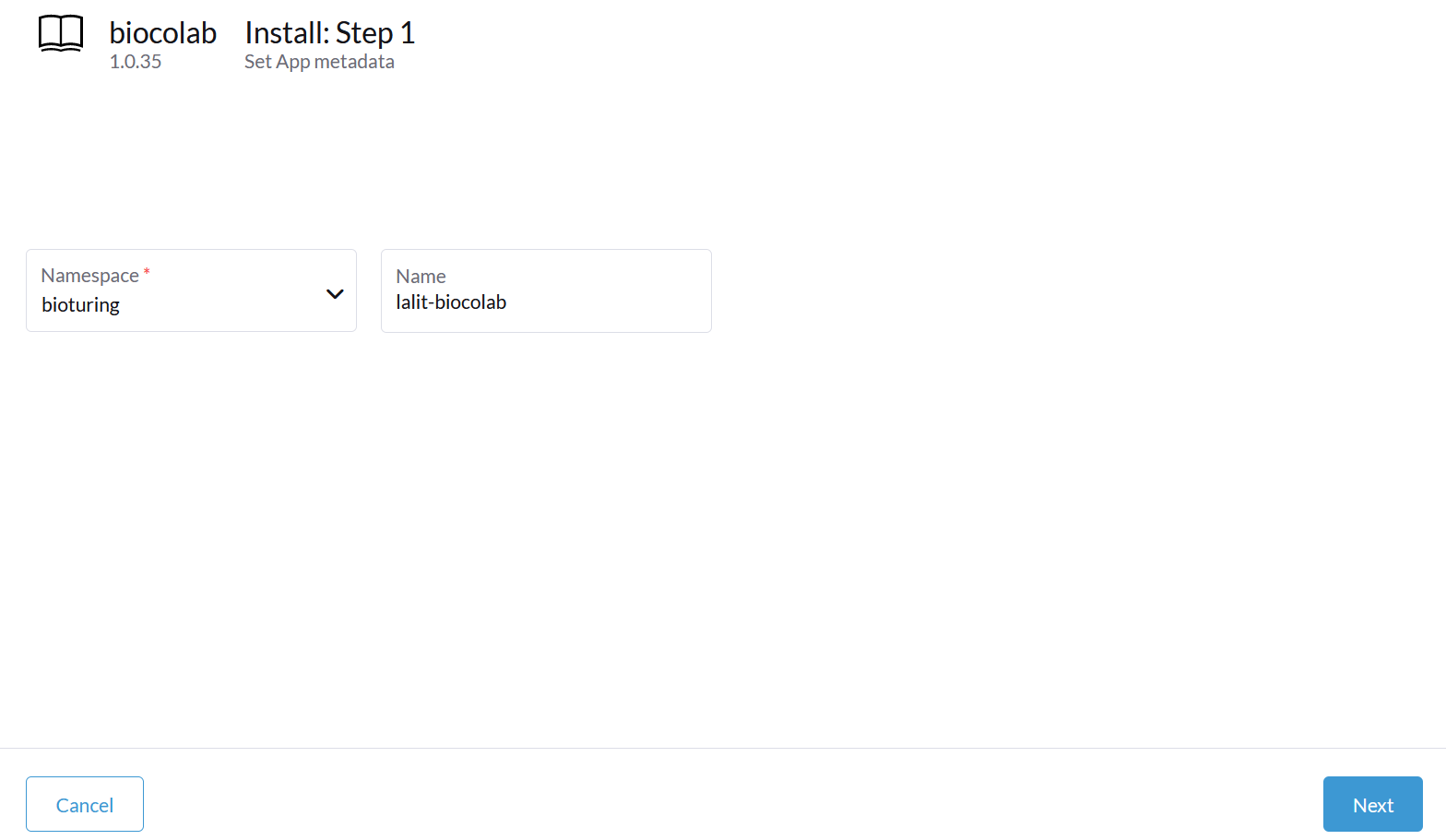
🔶 Verify Tag and repository

🔶 Update values:
# Change volume size according to your data.
# Update domain name and Colab token.
# Update credentials based on your choice.
# Click on install.
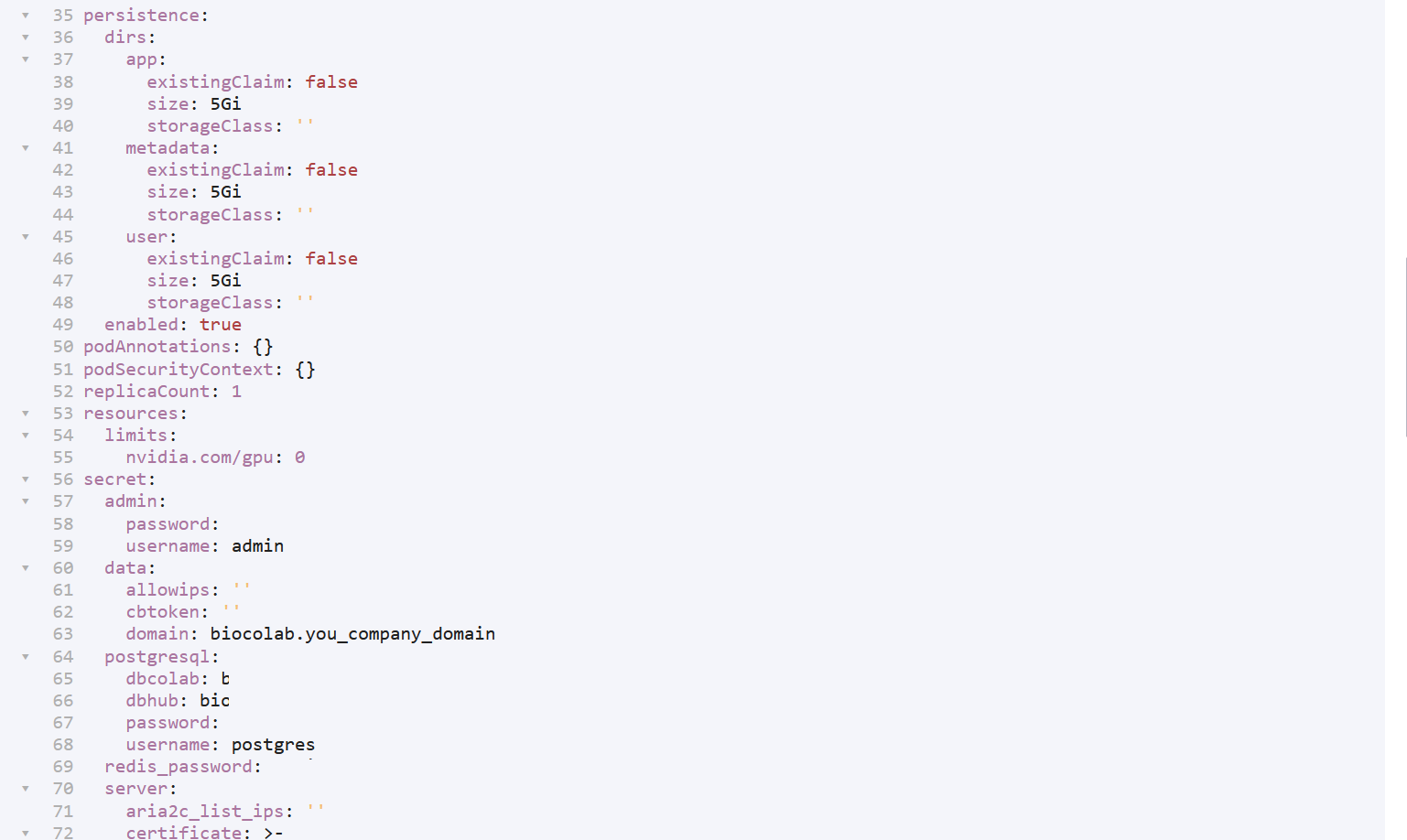
🔶 BioStudio will start installation based on helm chart.
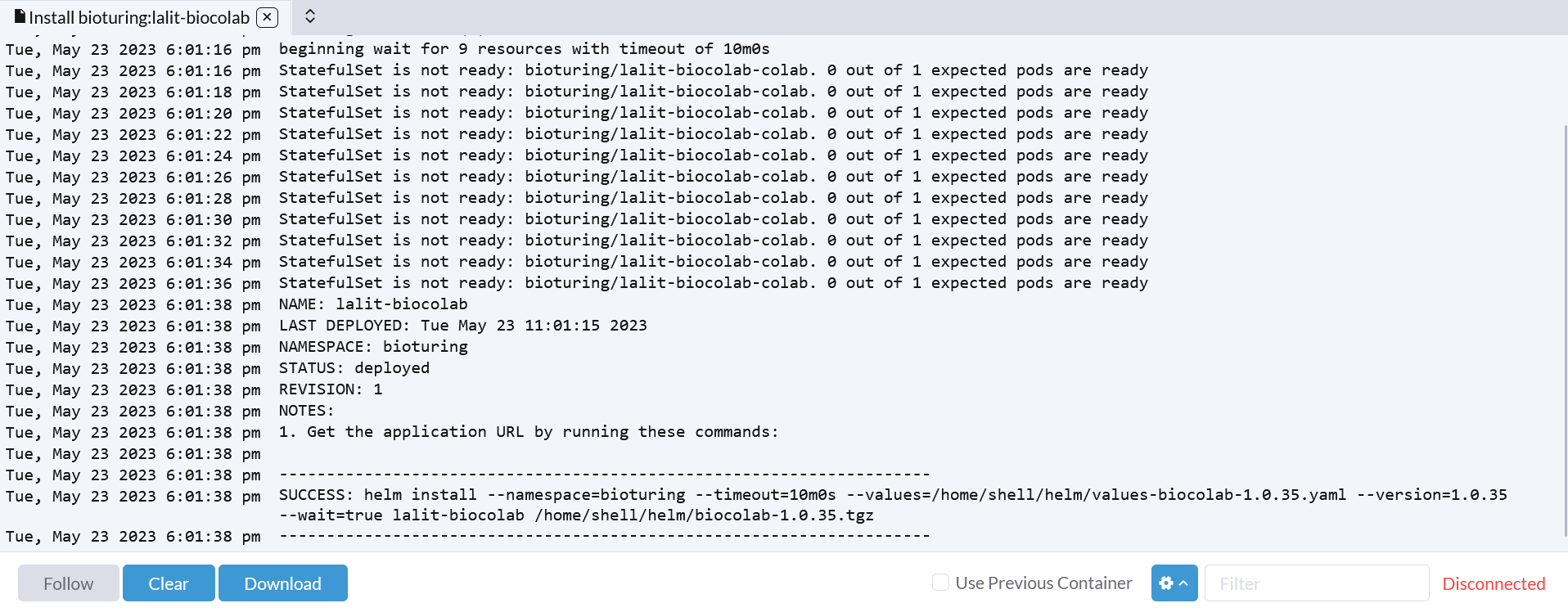
🔶 Verify statefulset.

🔶 Verify ingress.

🔶 Browse your domain.
🔸 Before access WORKSPACE. It is necessary to access dashboard and create machine and well as create volume to be mounted.
# Note: Kindly follow to create Machine and volume creation process in order to utilize our product.
# It is a necessary step.
# For any support and enquiry. Drop a mail to us support@bioturing.com

BioStudio Deployments on Kubernetes
¶Introduction
¶
BioStudio on K8s BioStudio software is compatible and easy to install on all kind of Operating System as well as on Docker, Kubernetes, Standalone machine, Cloud, On-premises, VM and all type of infrastructure platform. We are happy to inform that we are a leading brands company to provide all of our Softwares as a Service (SaaS). It is directly ready to use for end users.
🔔 Please contact 📧 [support@bioturing.com] in case you need any support or have any inquiry for us.
We are supporting all kind of service provider including on-premise.
BioStudio on K8s. Install Software on Kubernetes Clusters with the Helm.
AWS EKS Cluster ¶
🛂 Login to AWS console.
⭕ Serach for EKS in serach bar.
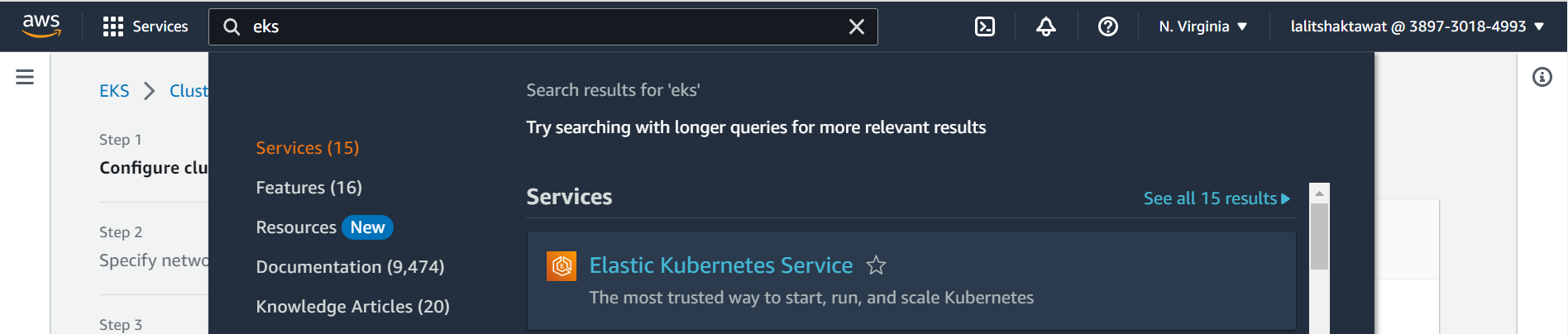
⭕ select create on Add Cluster.

⭕ Configure Cluster.
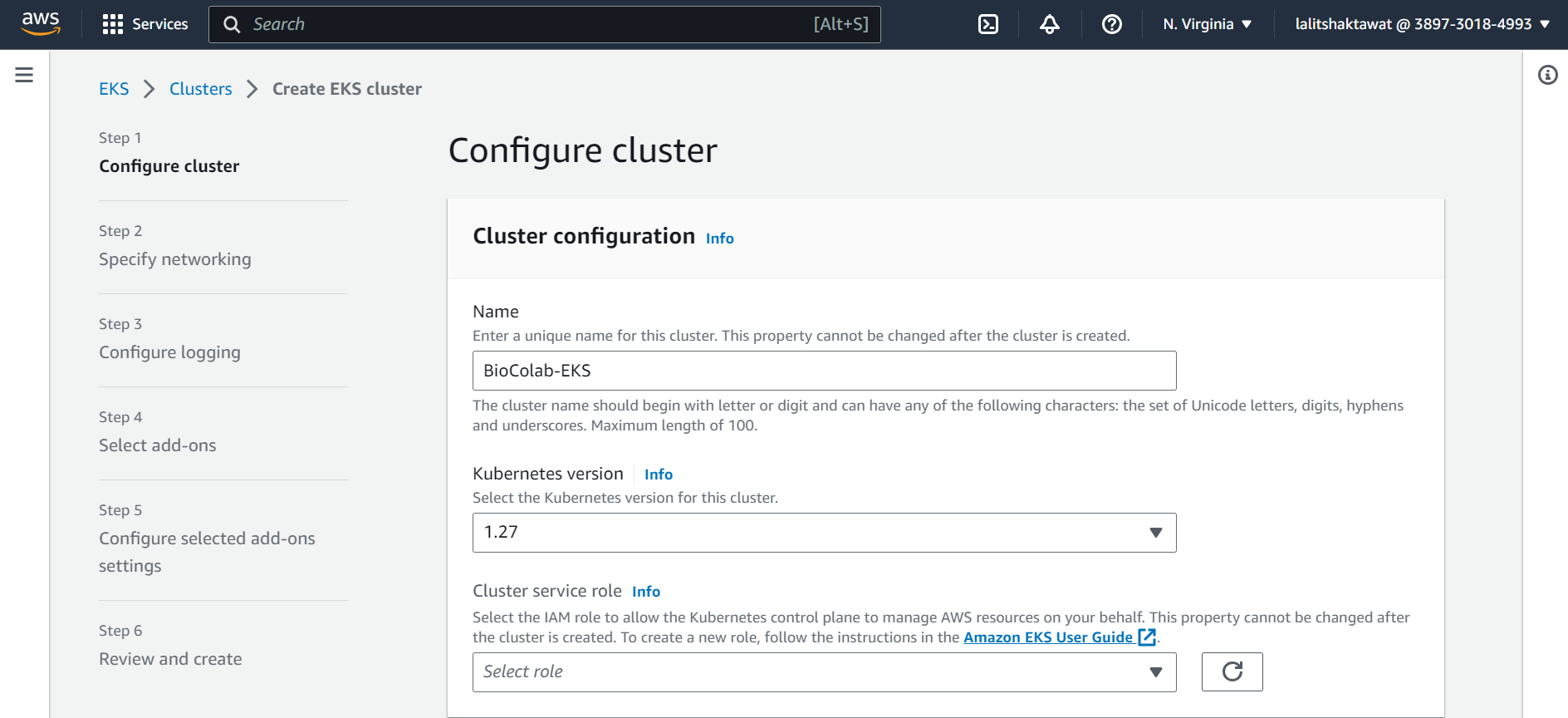
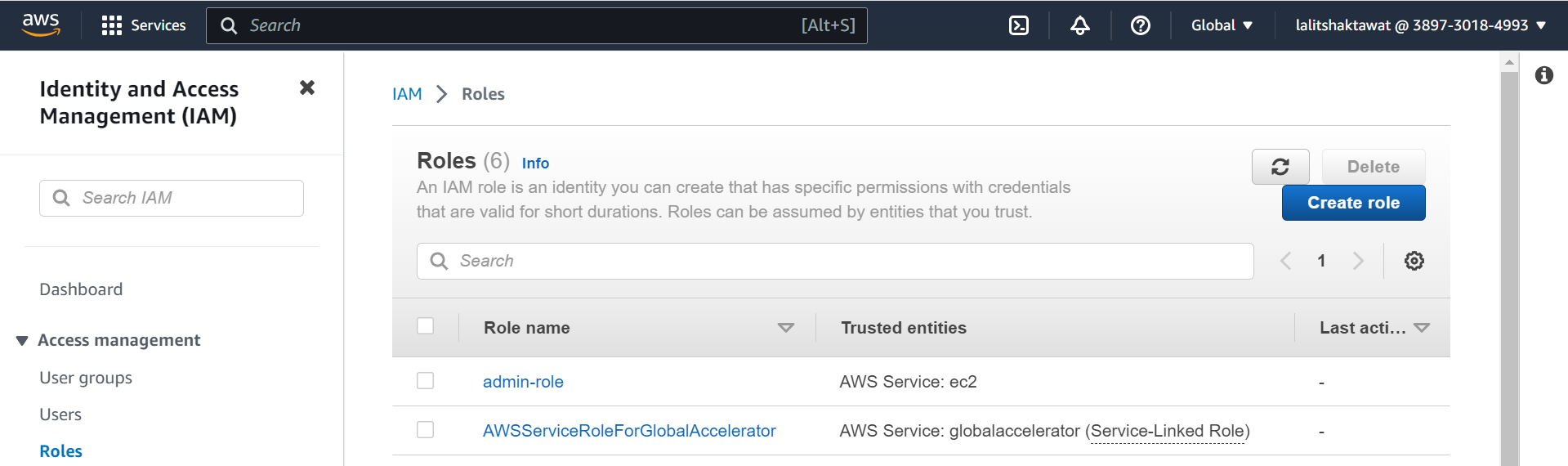
⭕ Create Cluster Service role.
role --> Create role
⭕ Select AWS service and EKS on use case.
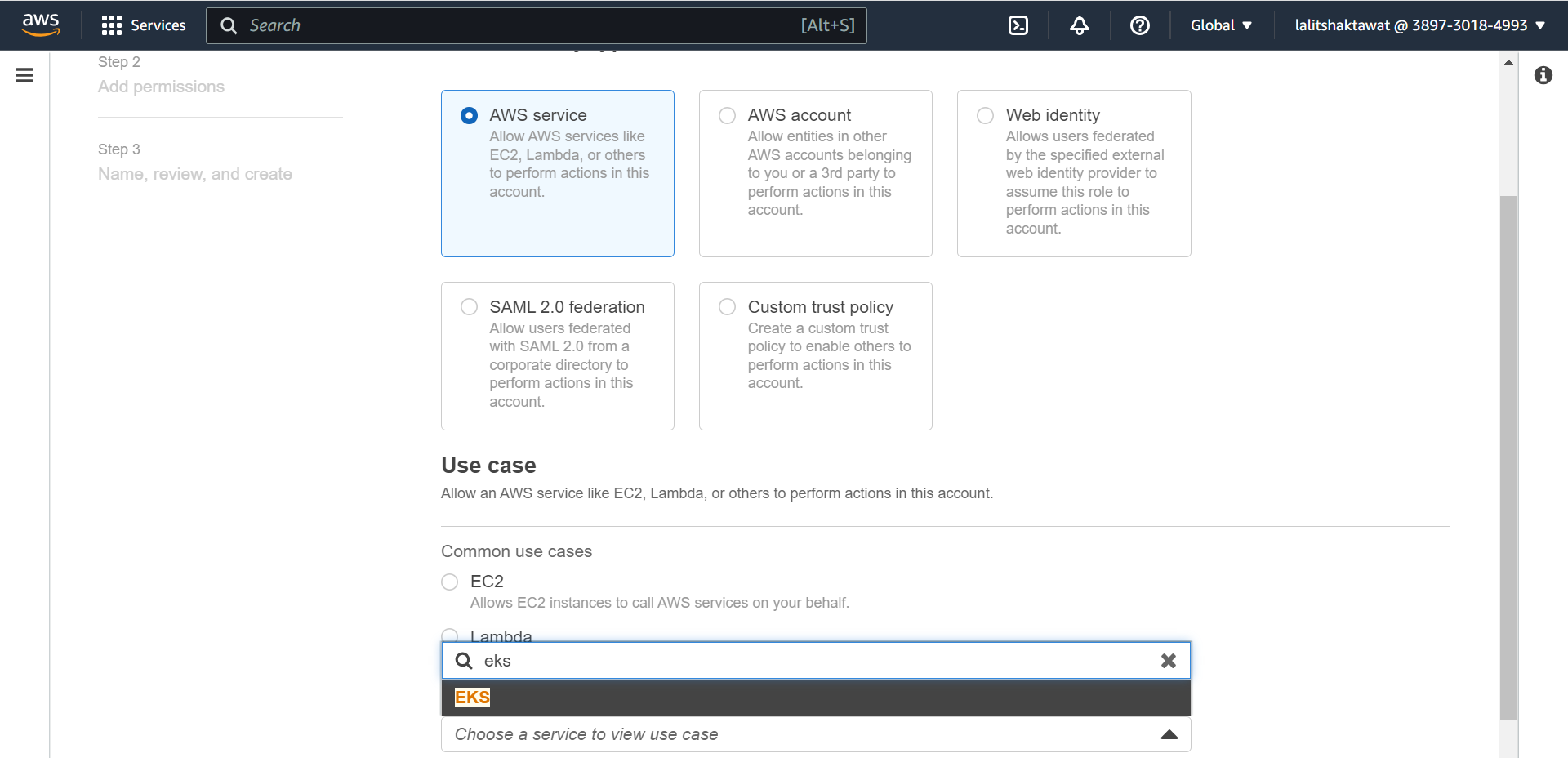
⭕ Select EKS -Cluster.
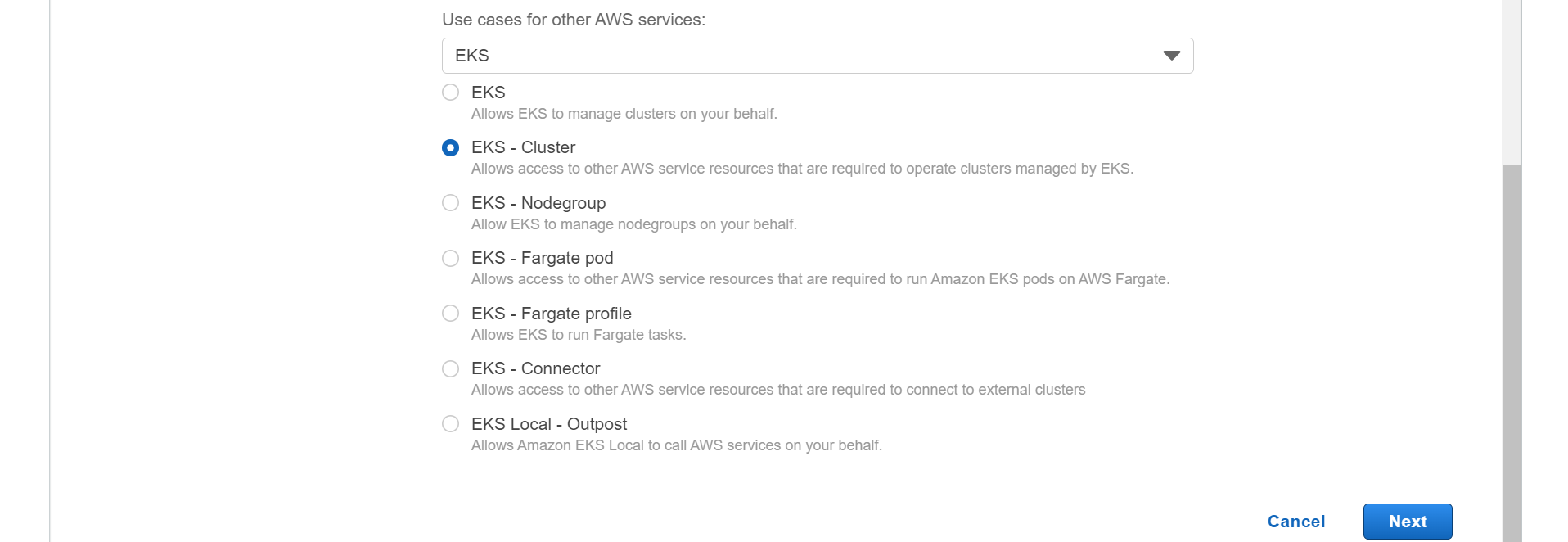
⭕ Add permission.
AmzonEKSClusterPolicy
➡️ click on next.
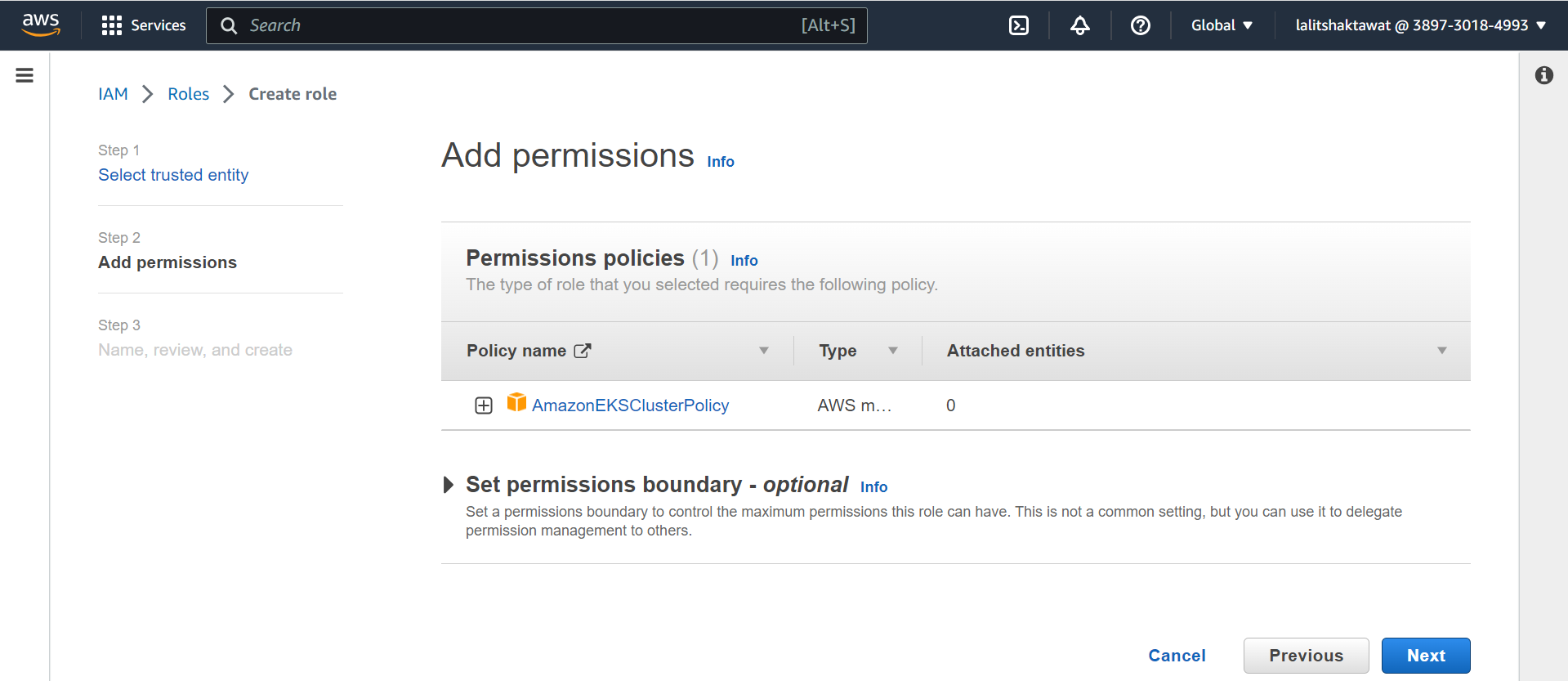
⭕ Fill Role detail.
➡️ Create role.
⭕ Role created .
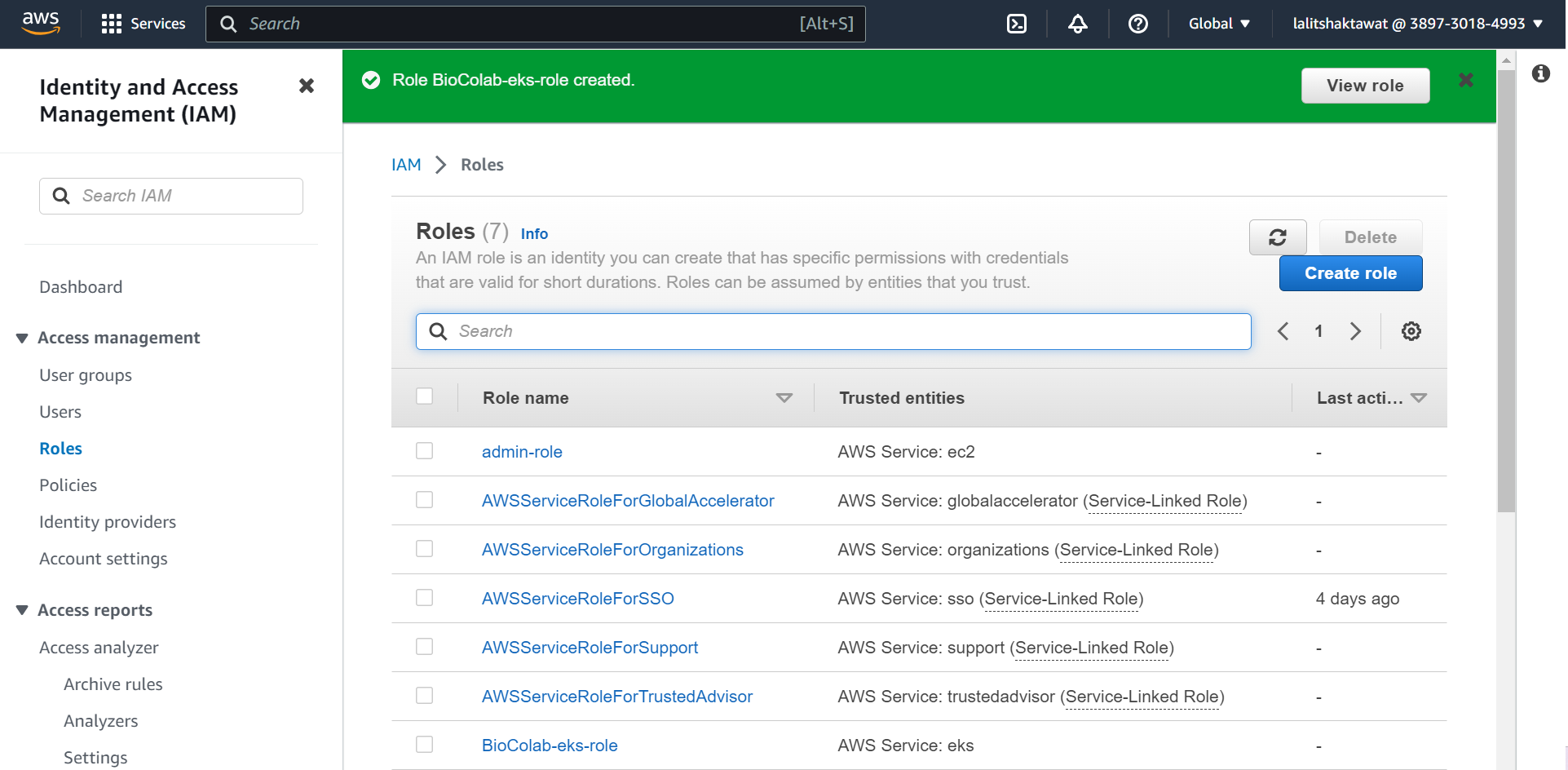
⭕ Select appropriate role that we created earlier..
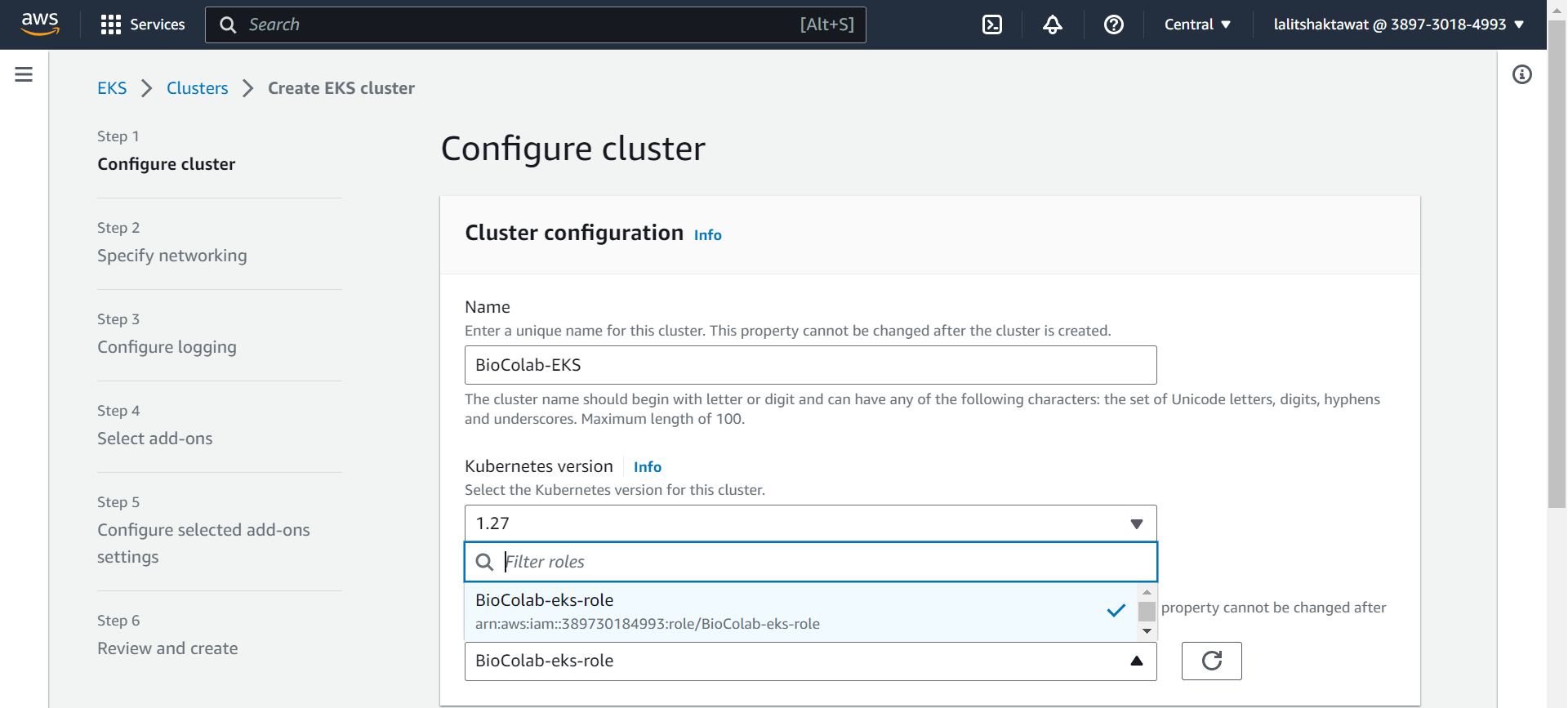
⭕ provide appropriate tag.
➡️ click on next.
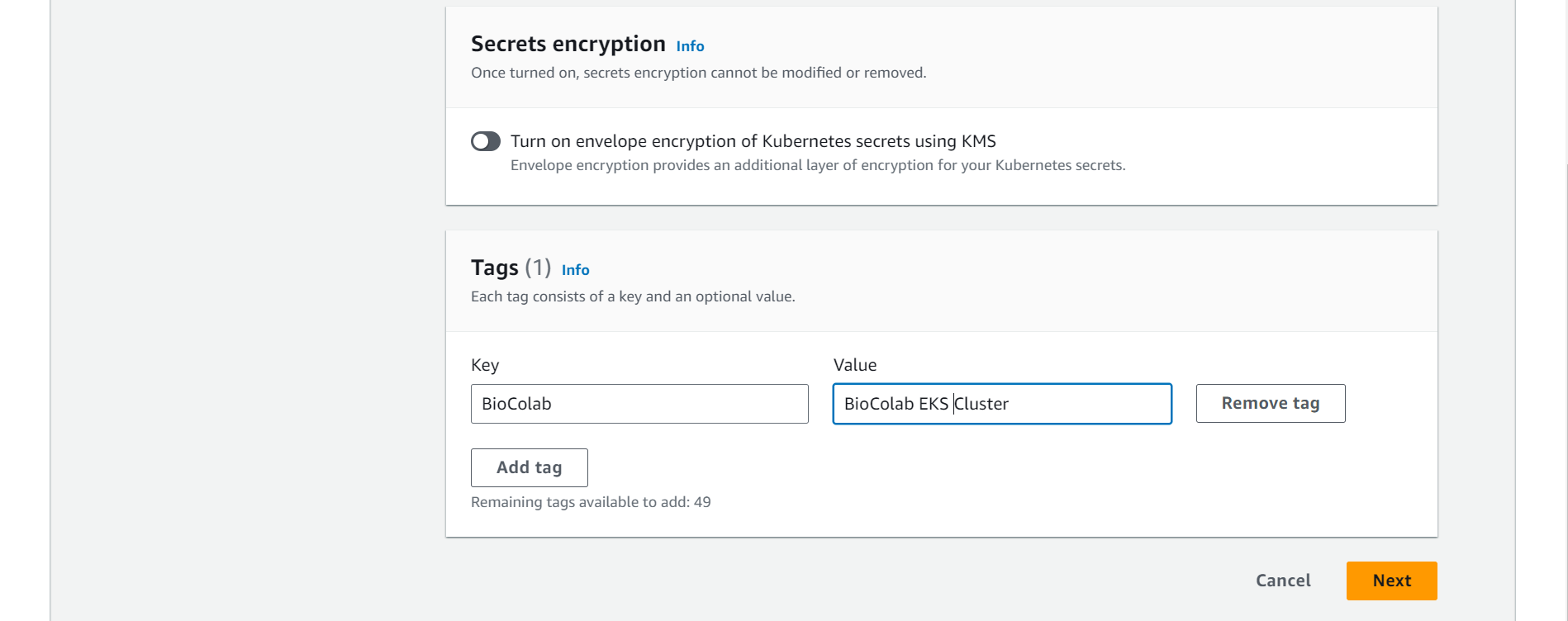
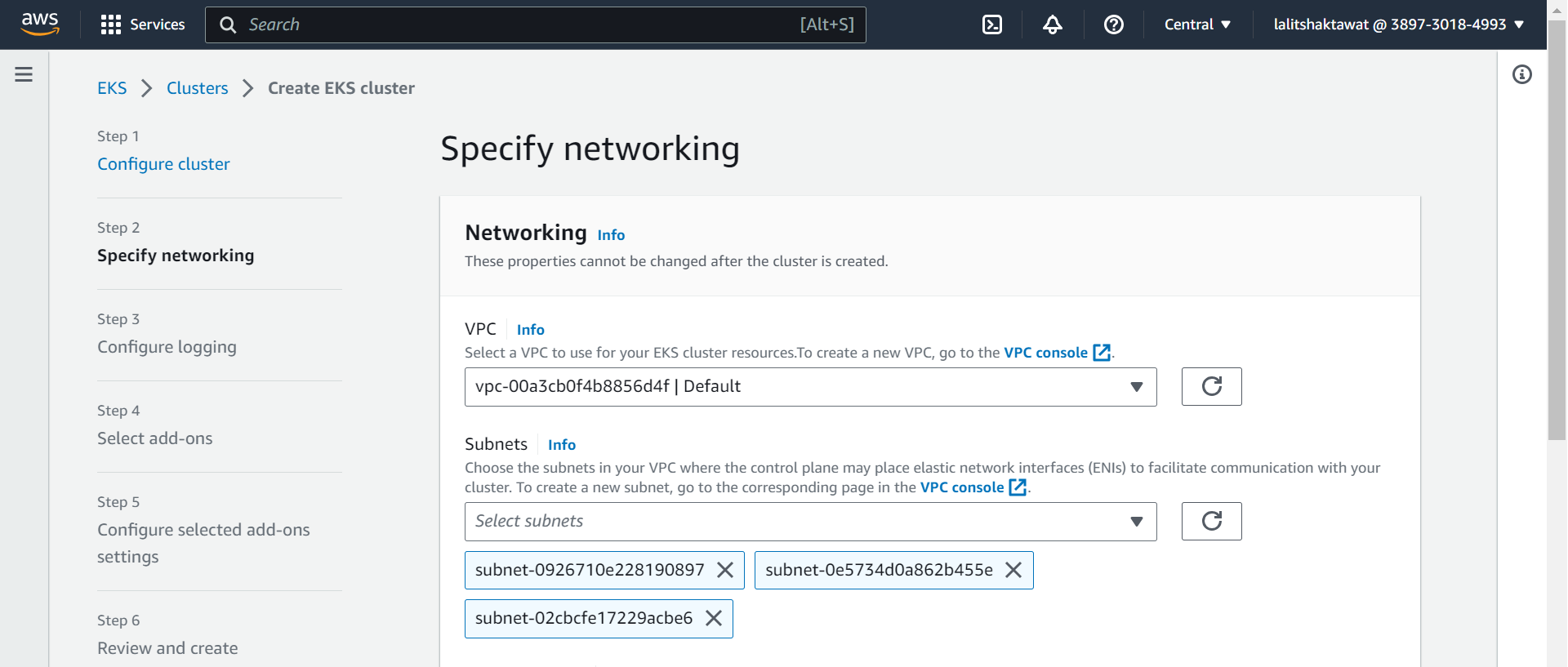
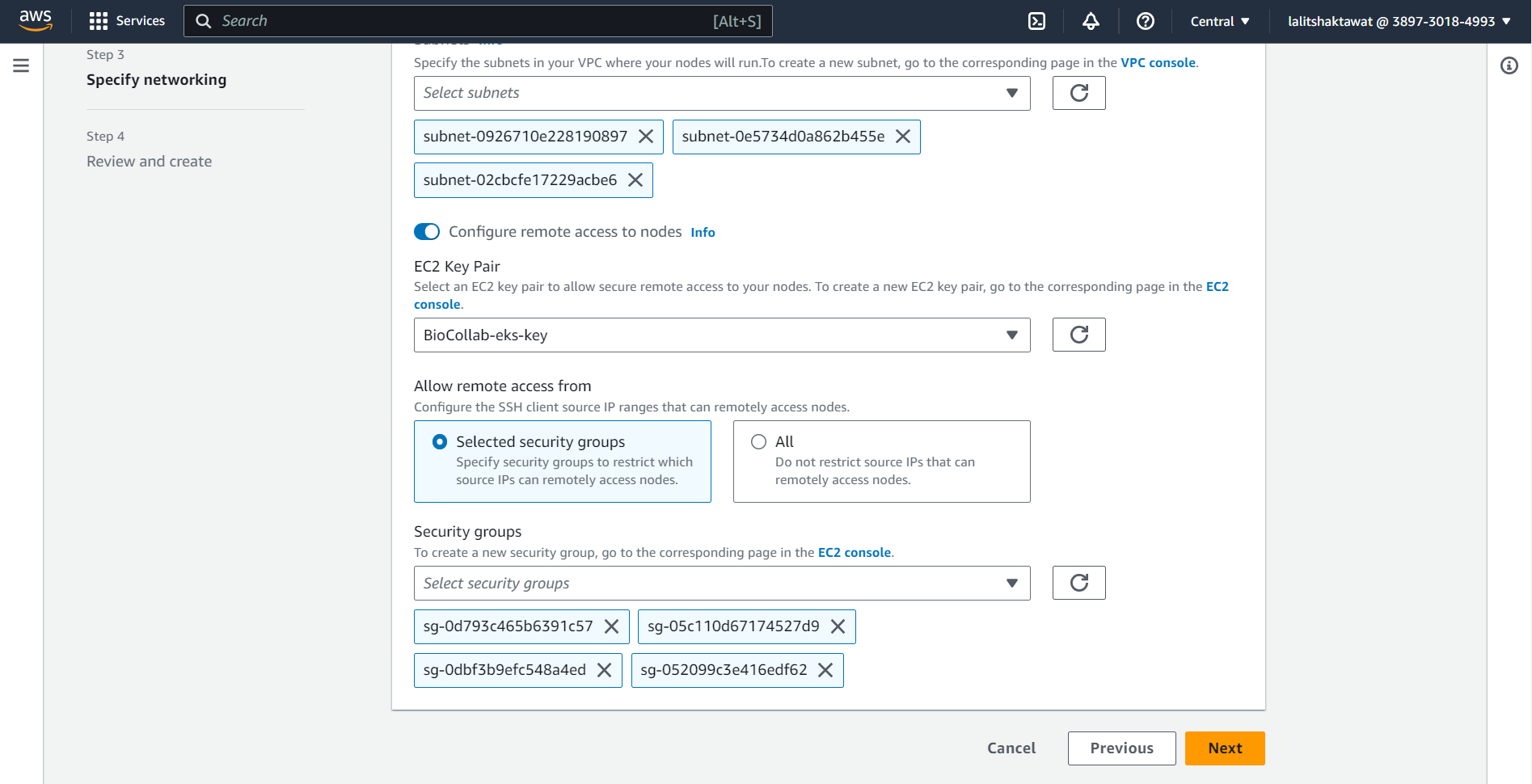
⭕ Specify networking. VPC and Networking.
🔗 Amazon EKS VPC and subnet requirements and considerations
⭕ Select Security Group.
➡️ click on next.
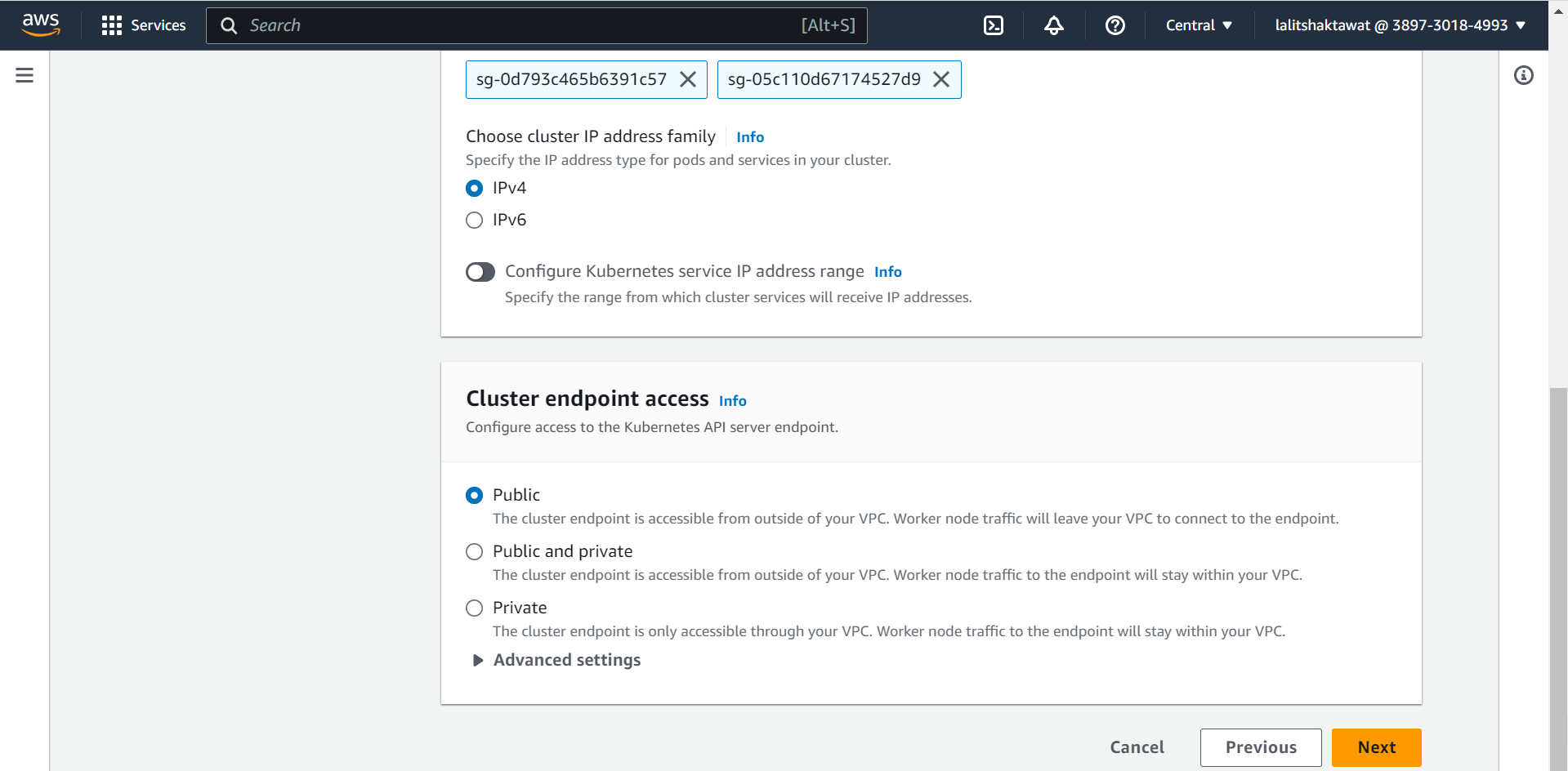
⭕ Select Configuring logging.
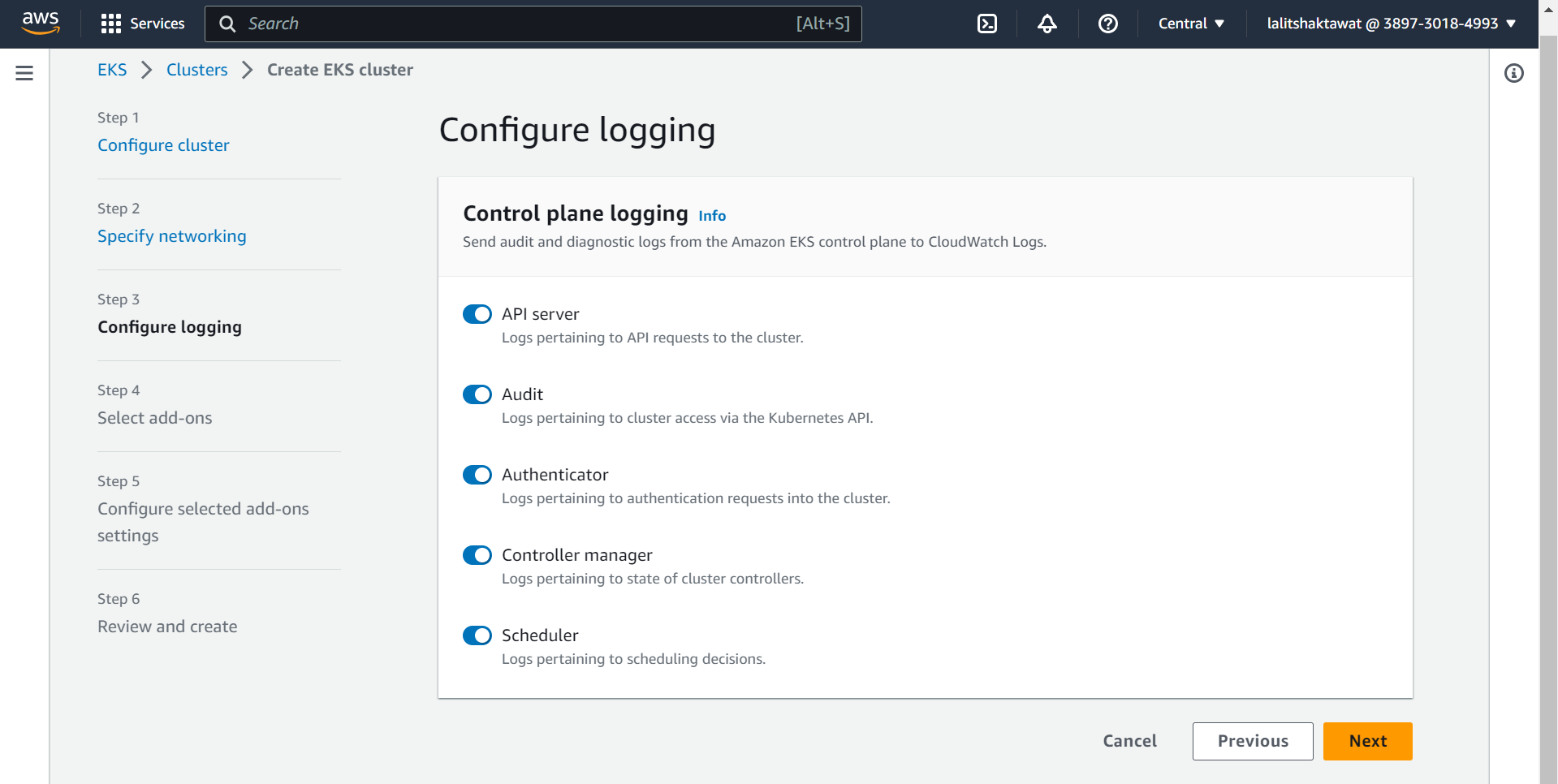
⭕ Select Add-ons.
➡️ click on next.
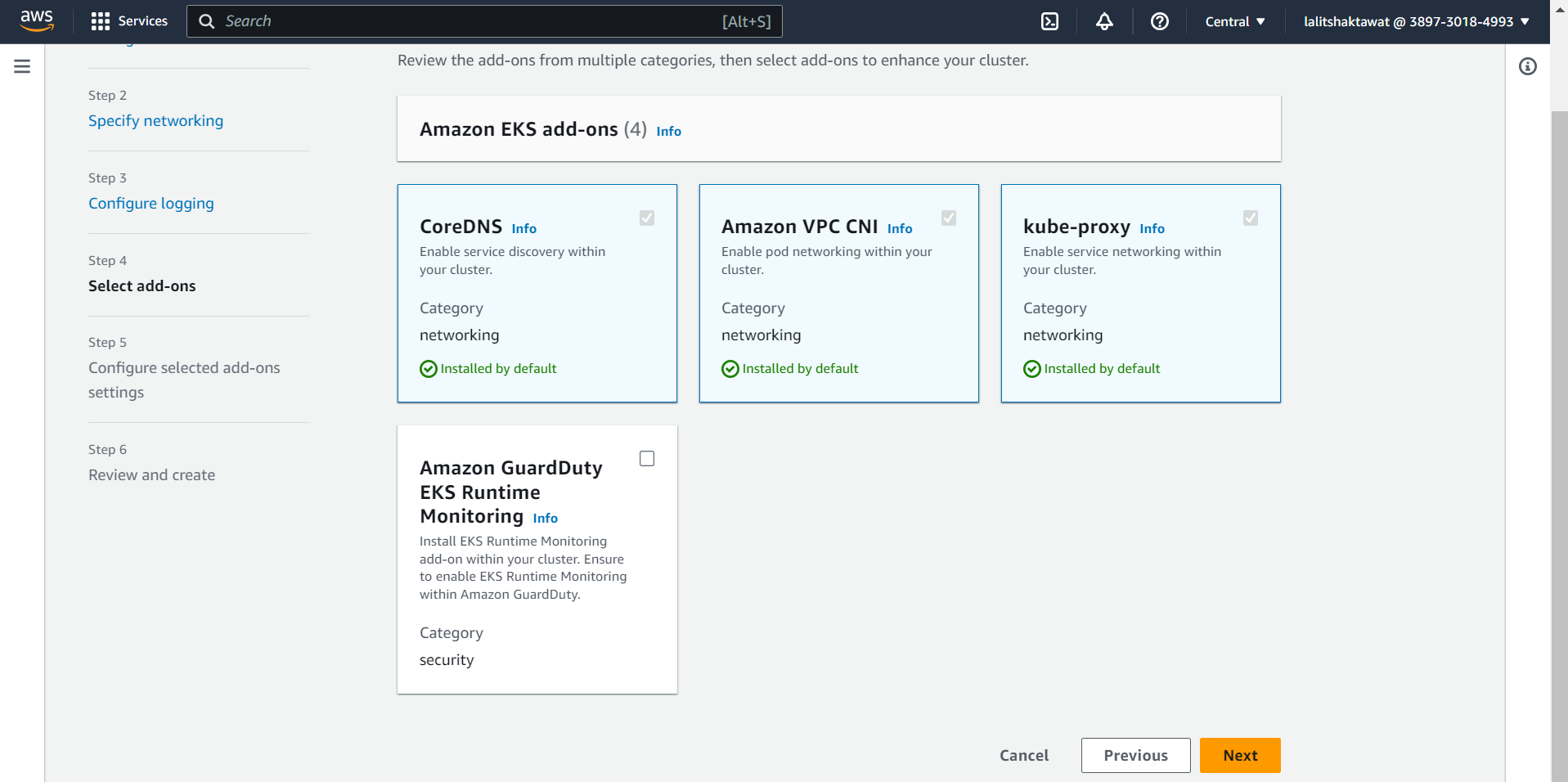
➡️ Select Version of Add-ons and click on next.
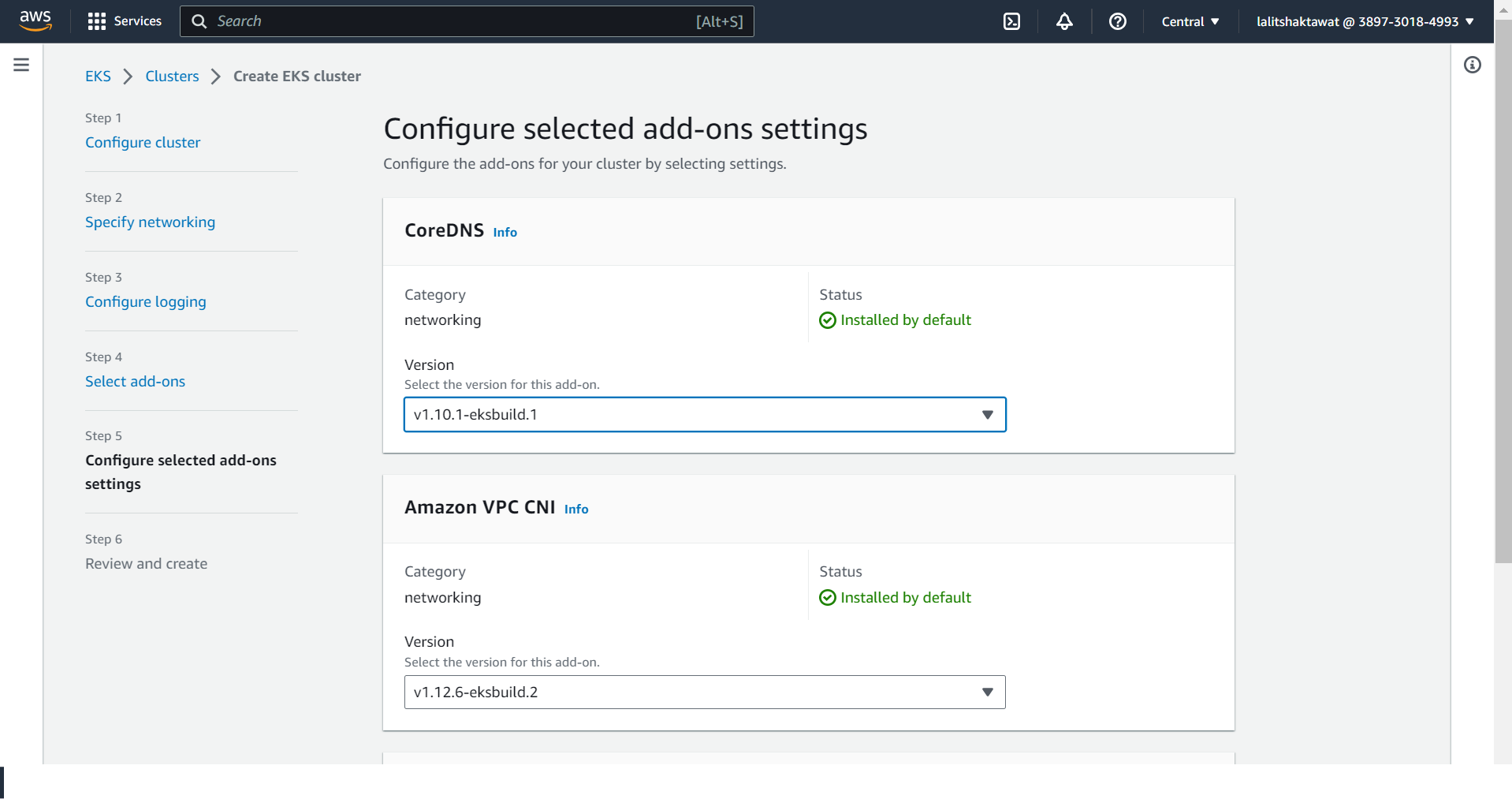
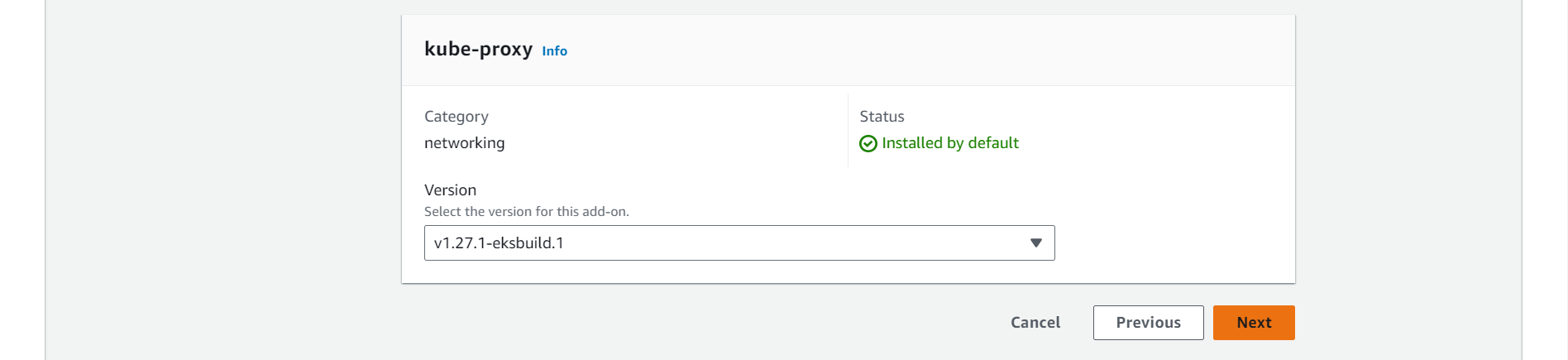
⭕ Click on Create.
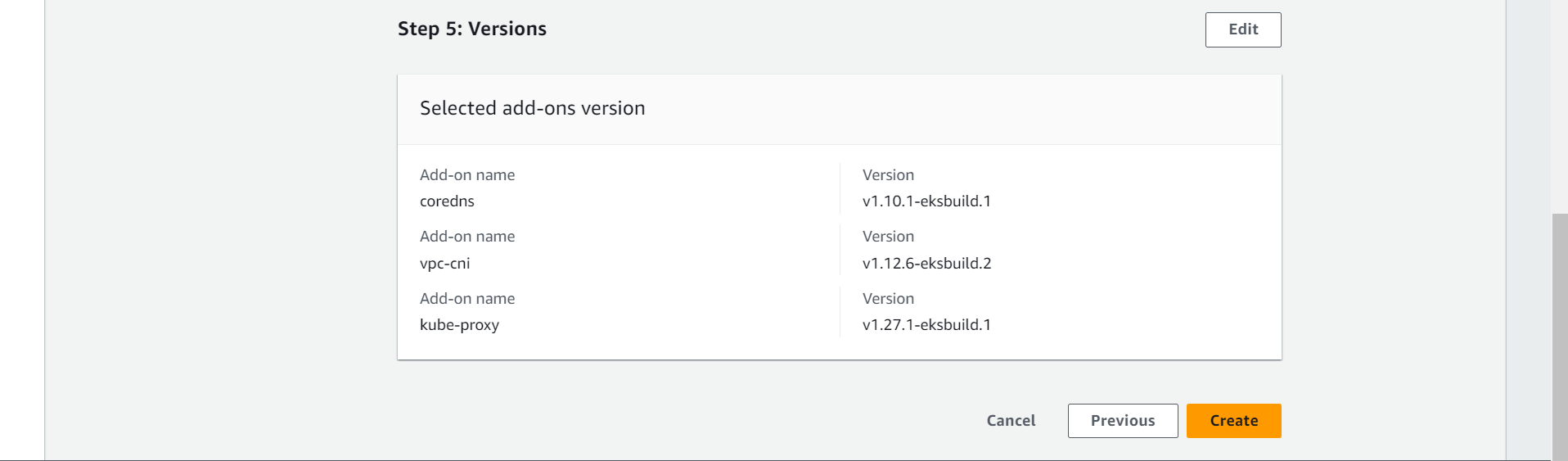
😊 You are done to create cluster. Wait for a while to setup this.
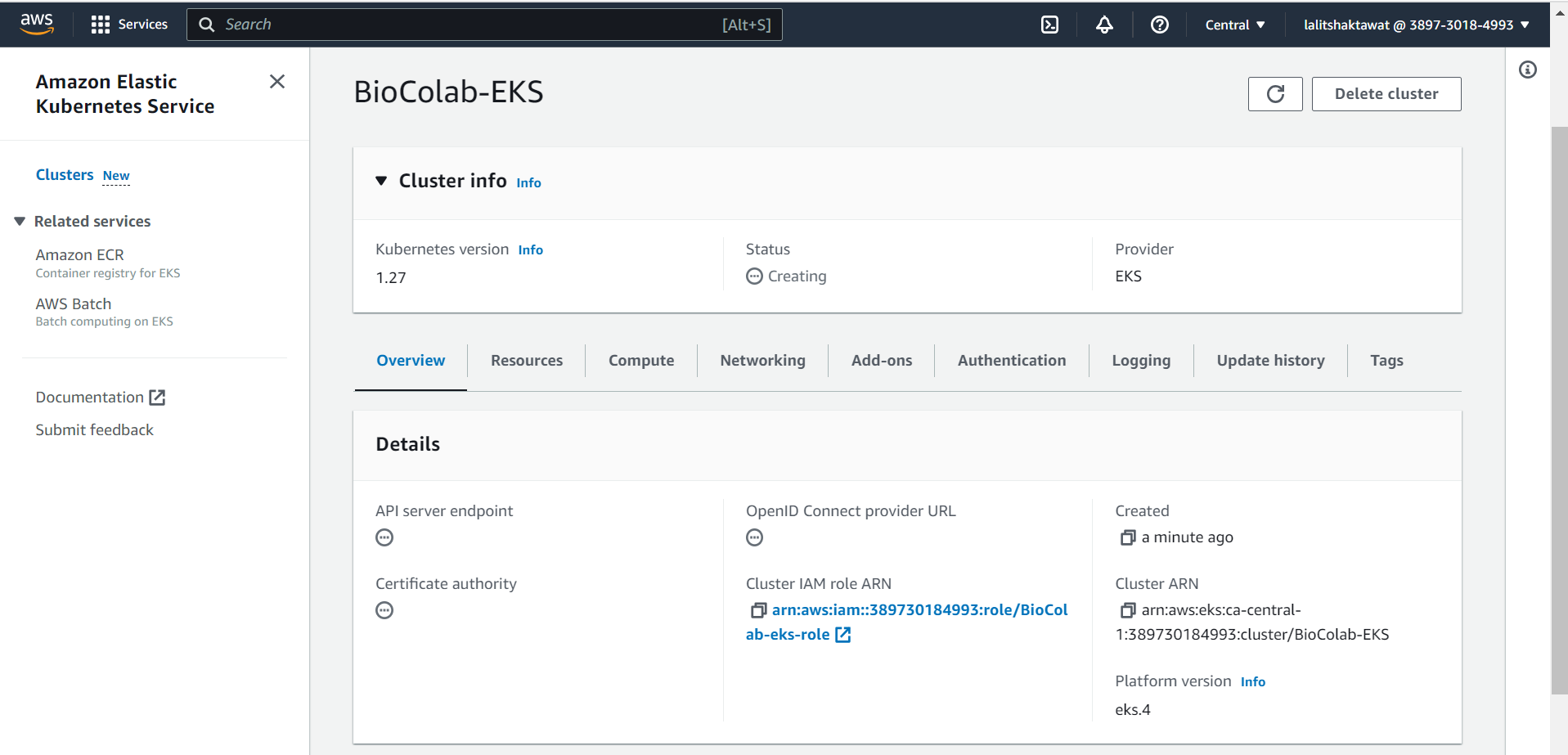
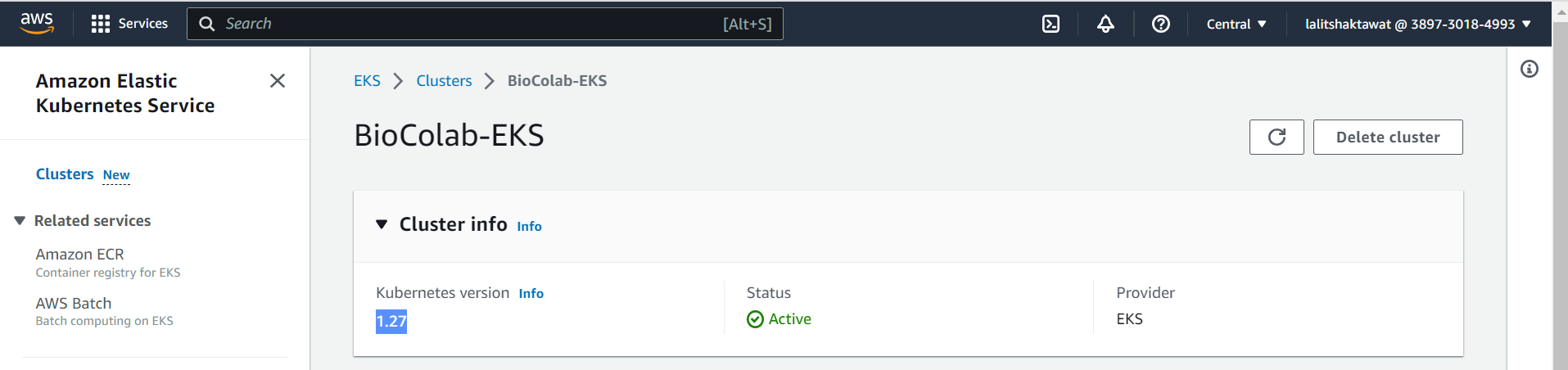
🎁 Kubernetes Cluster is Active now.
🔗 AWS CLI
Kindly select the OS version installation according to your Operating System.
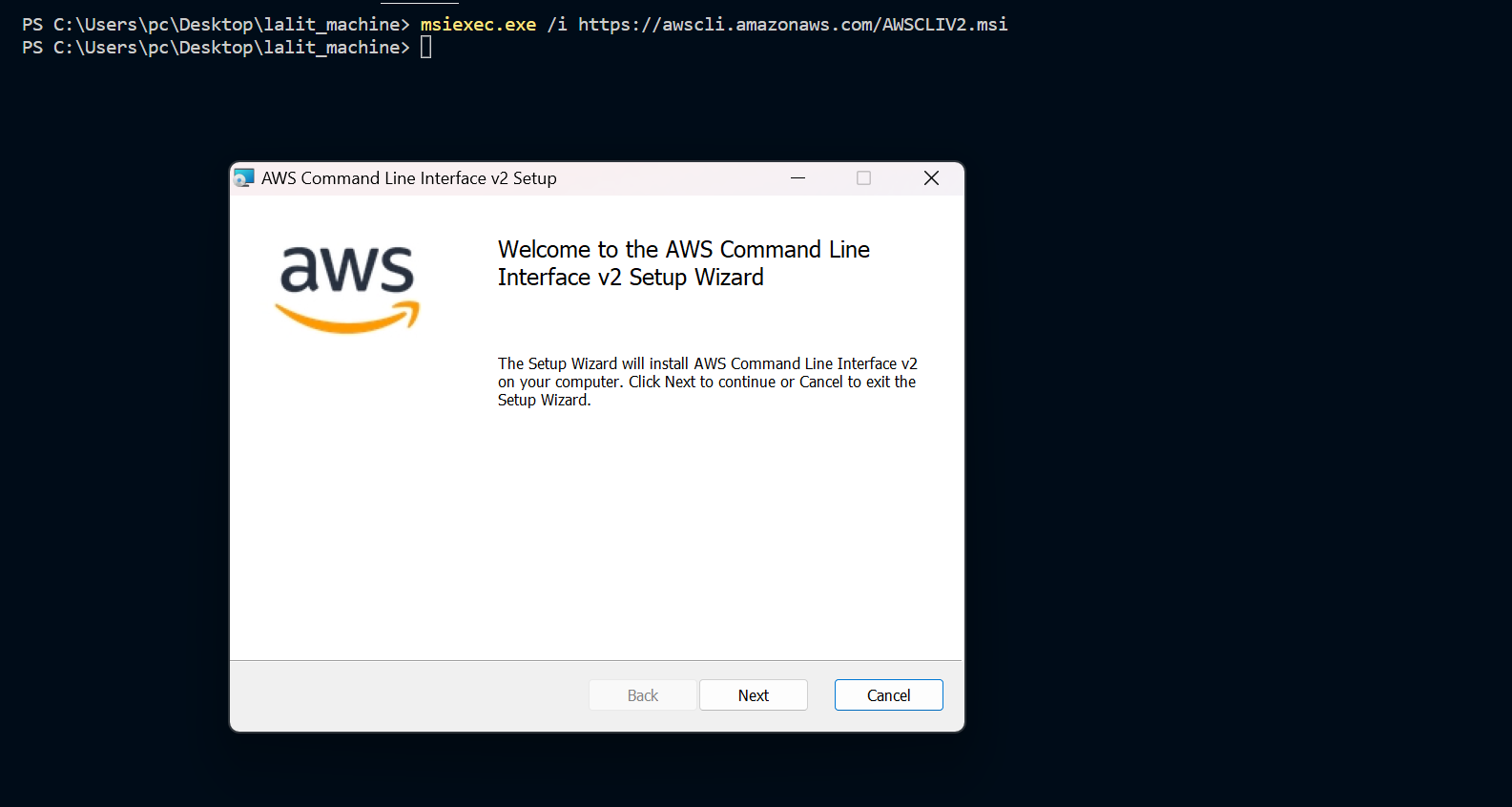
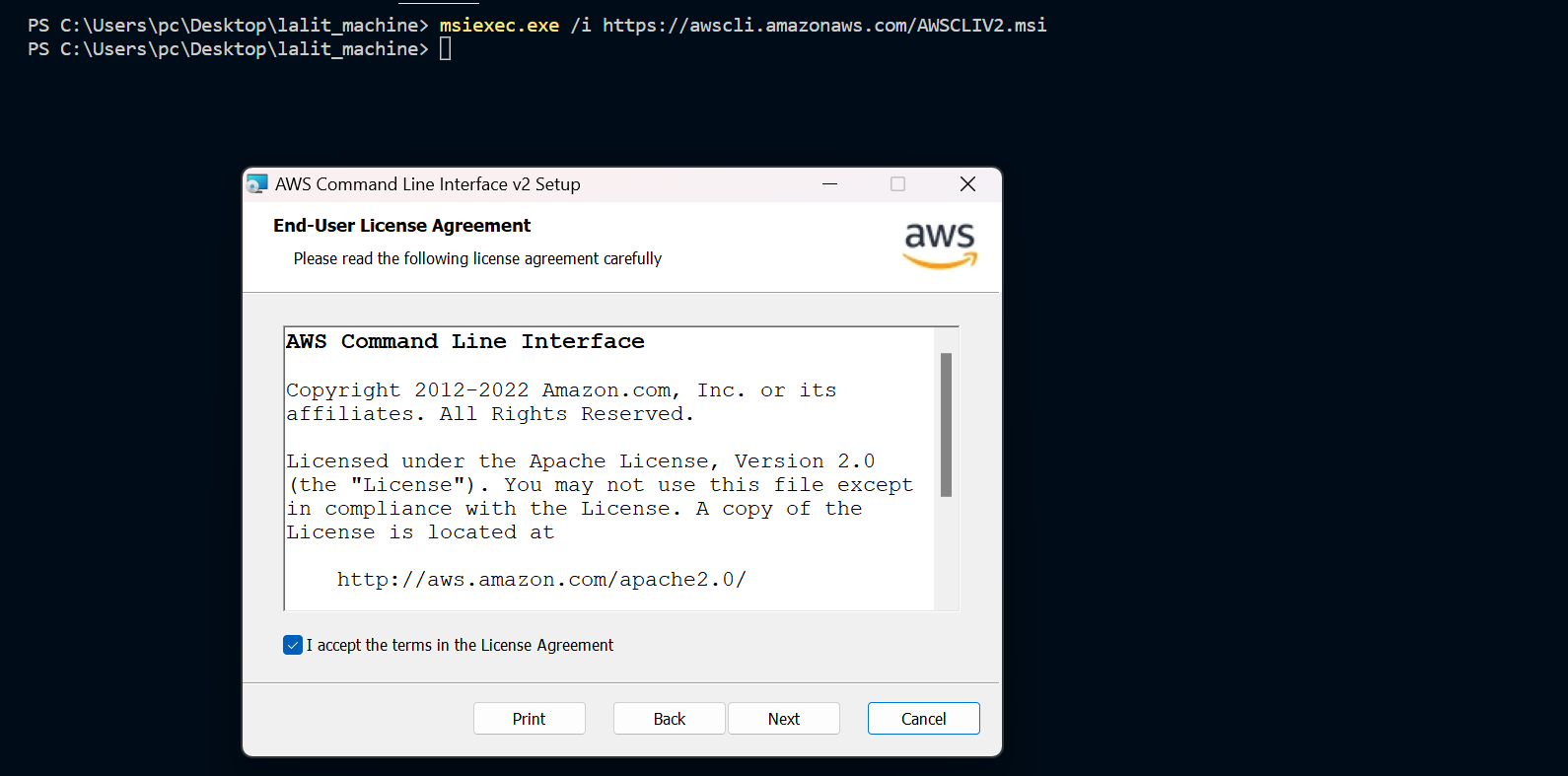
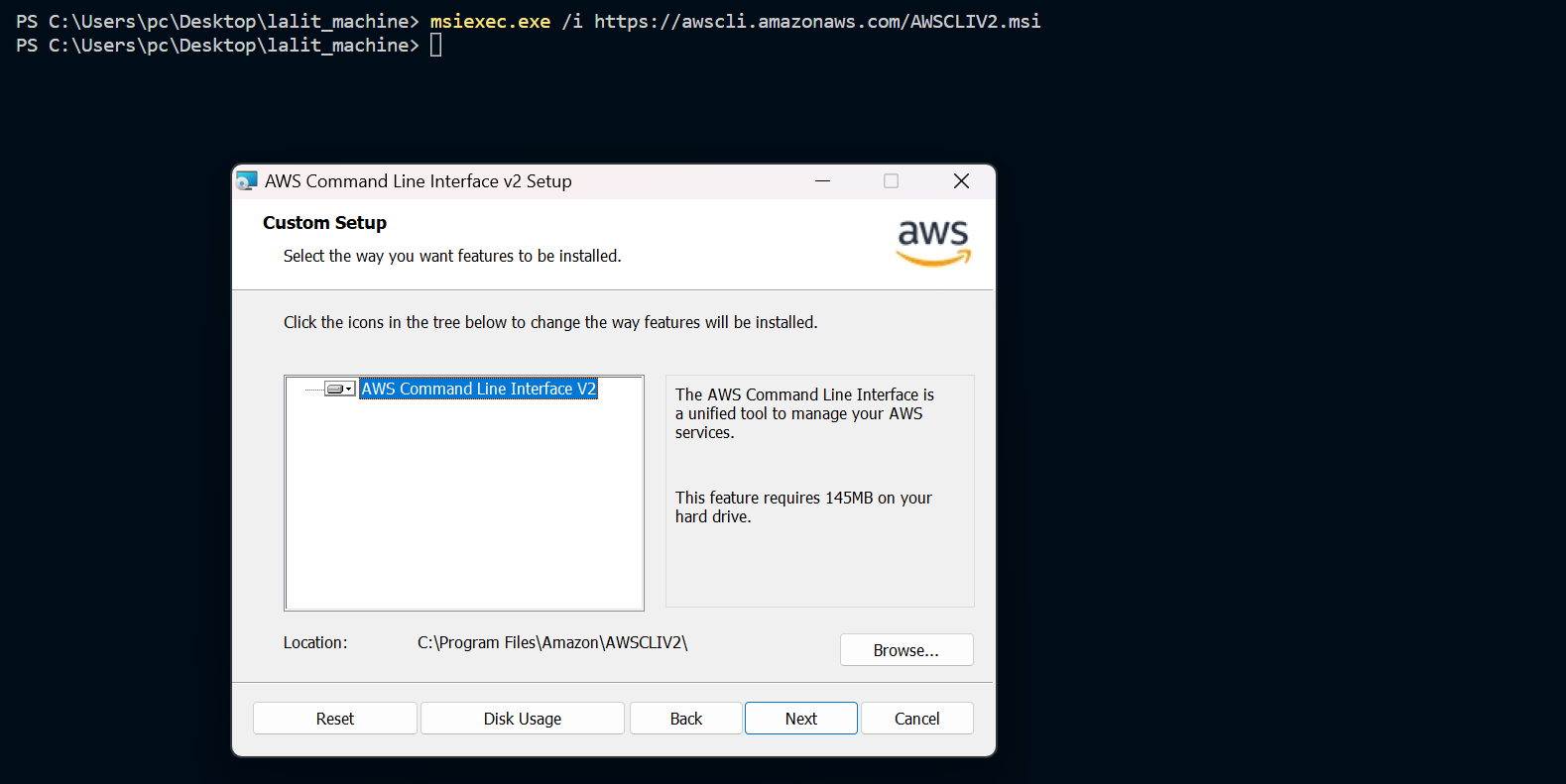
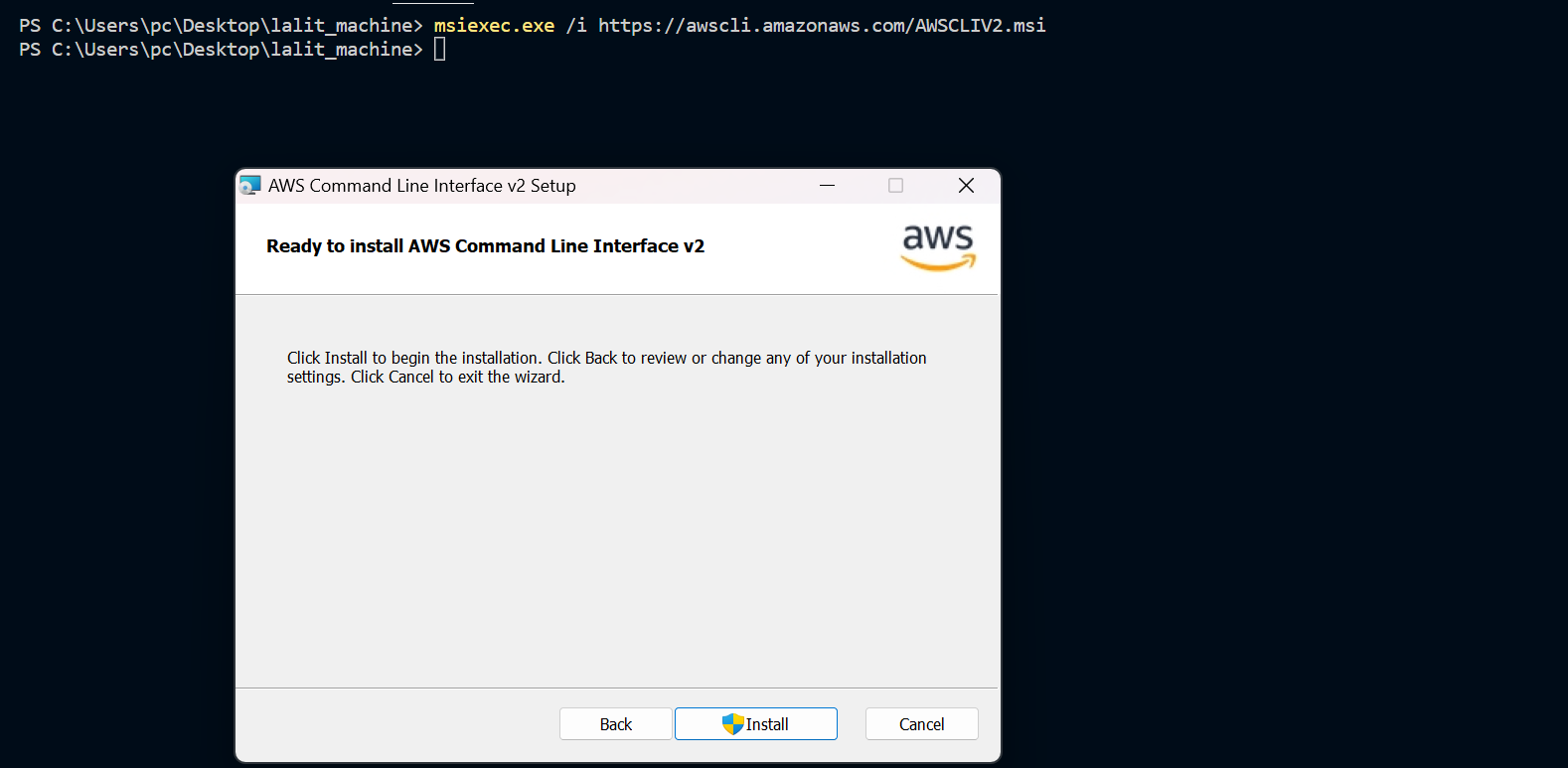
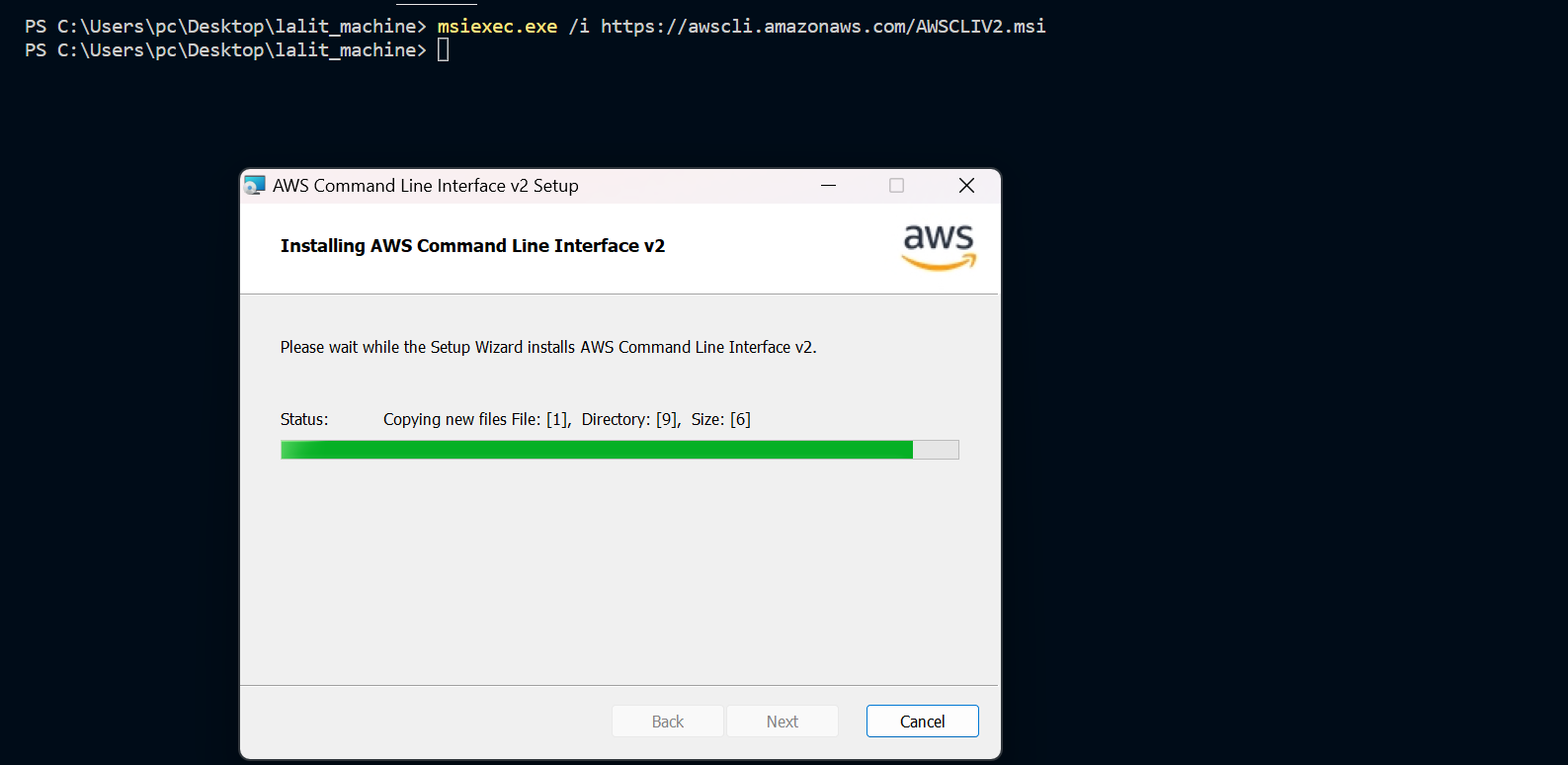
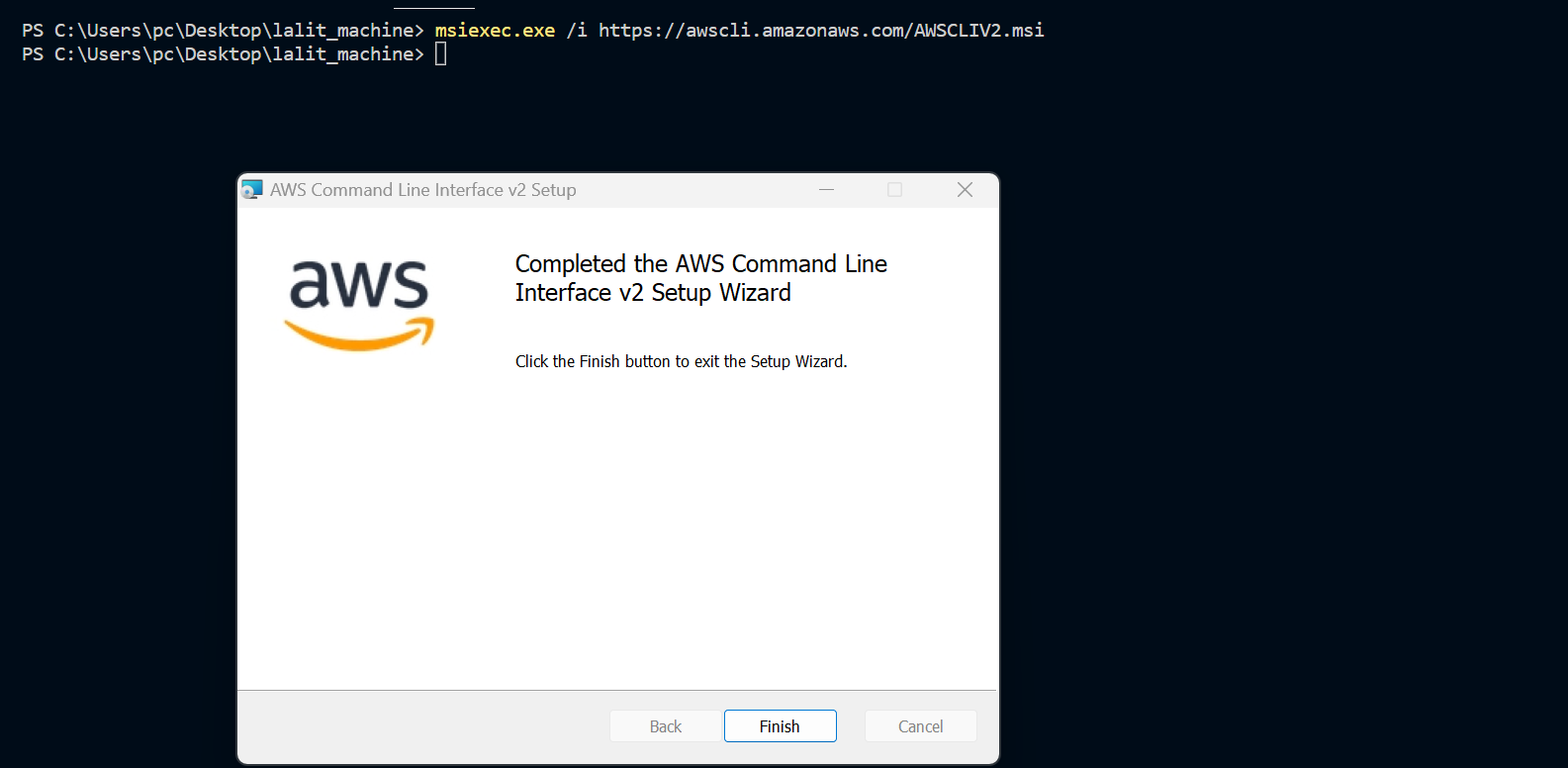
⭕ Test AWS cli


⭕ Installing Kubectl command utility.
There are many ways to install Kubectl command utility too. Here we are using window machine, So just to prefer Choco.
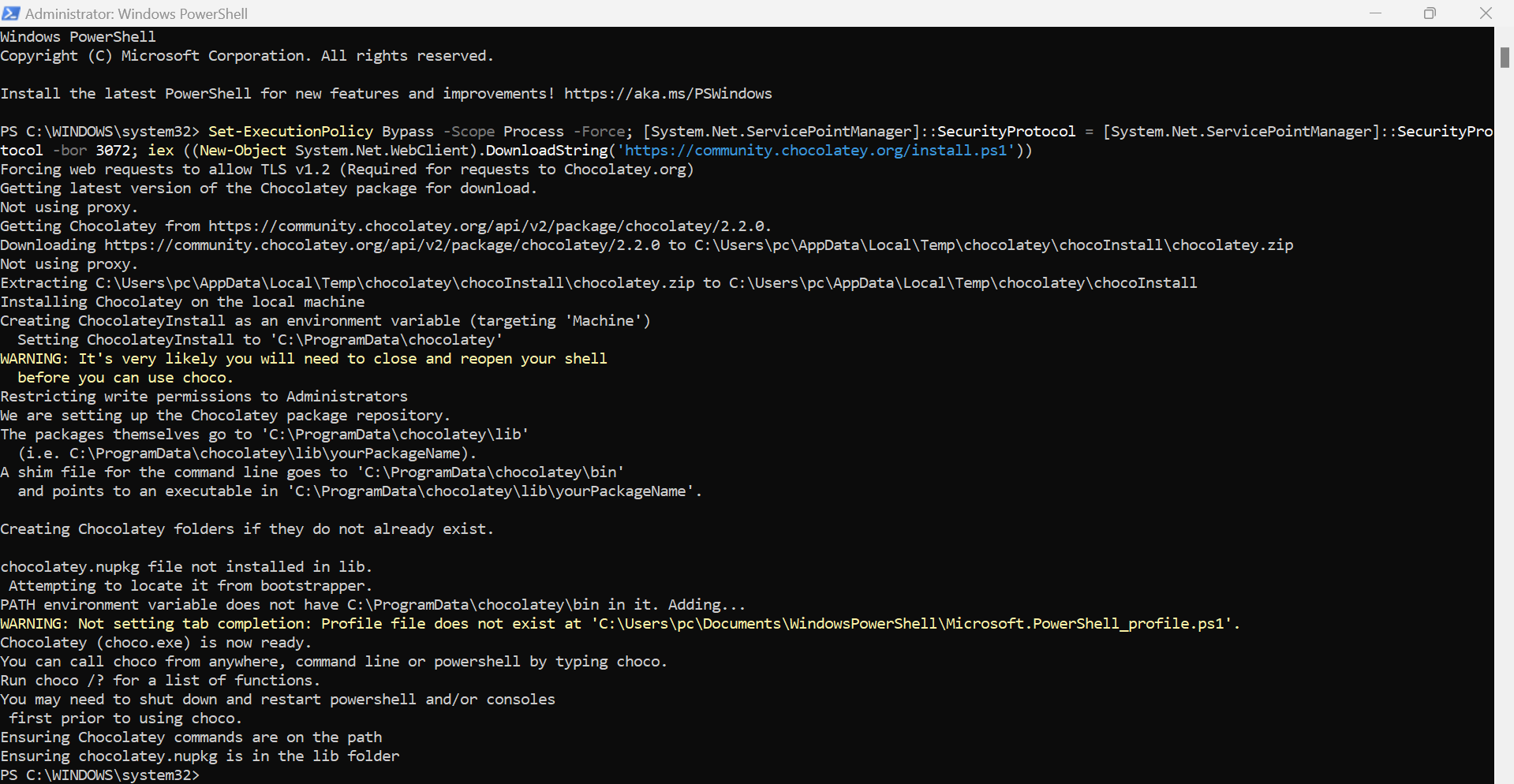
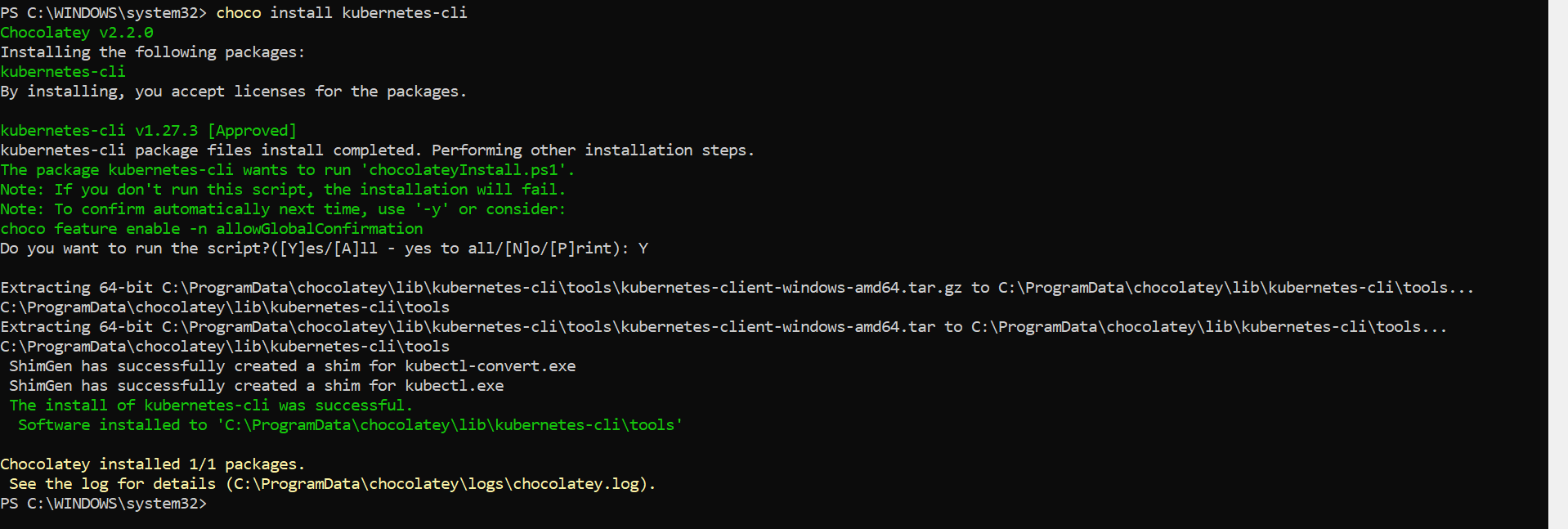
⭕ Adding worker node to Kubernete cluster.
EKS --> Compute --> Add node group.
In order to add node to the cluster. We need to add node group. Each node group can content different kind of node ( compute resources ). We can add many node group based on requirements.
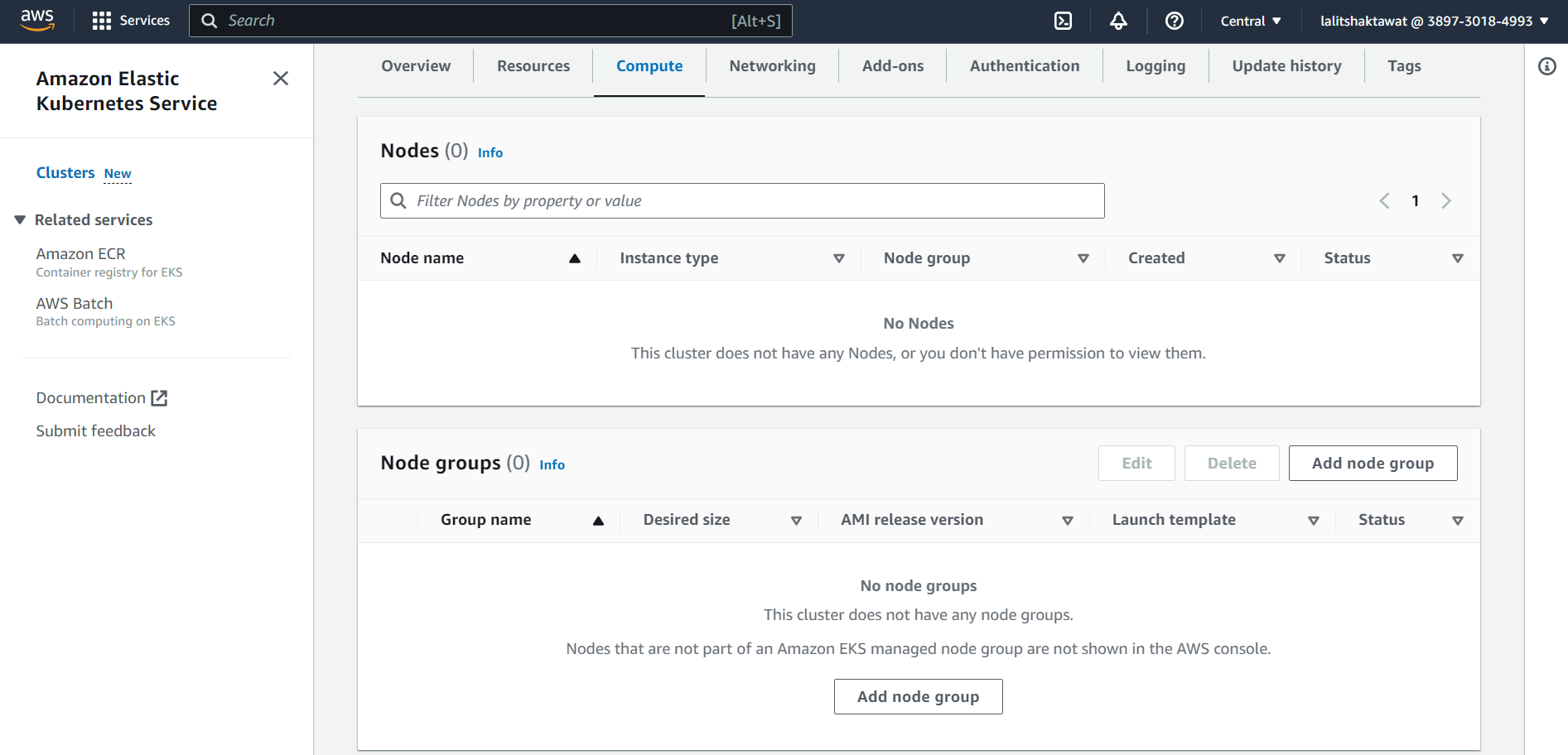
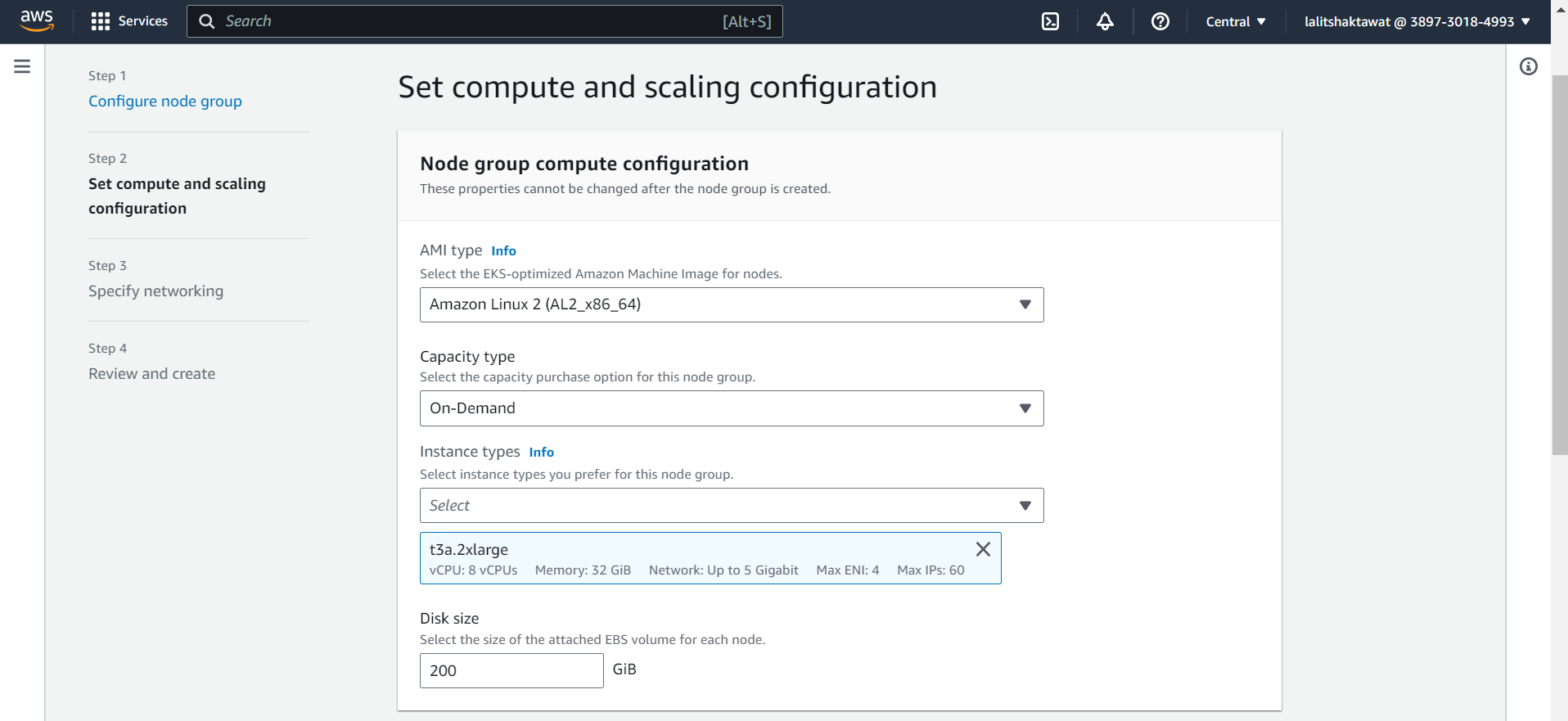
⭕ Configure node group.
In node group configuration, We need to provide node name as well as must create Node IAM role. to handle worker node. Once click on IAM console. A new window will open to create new IAM role.
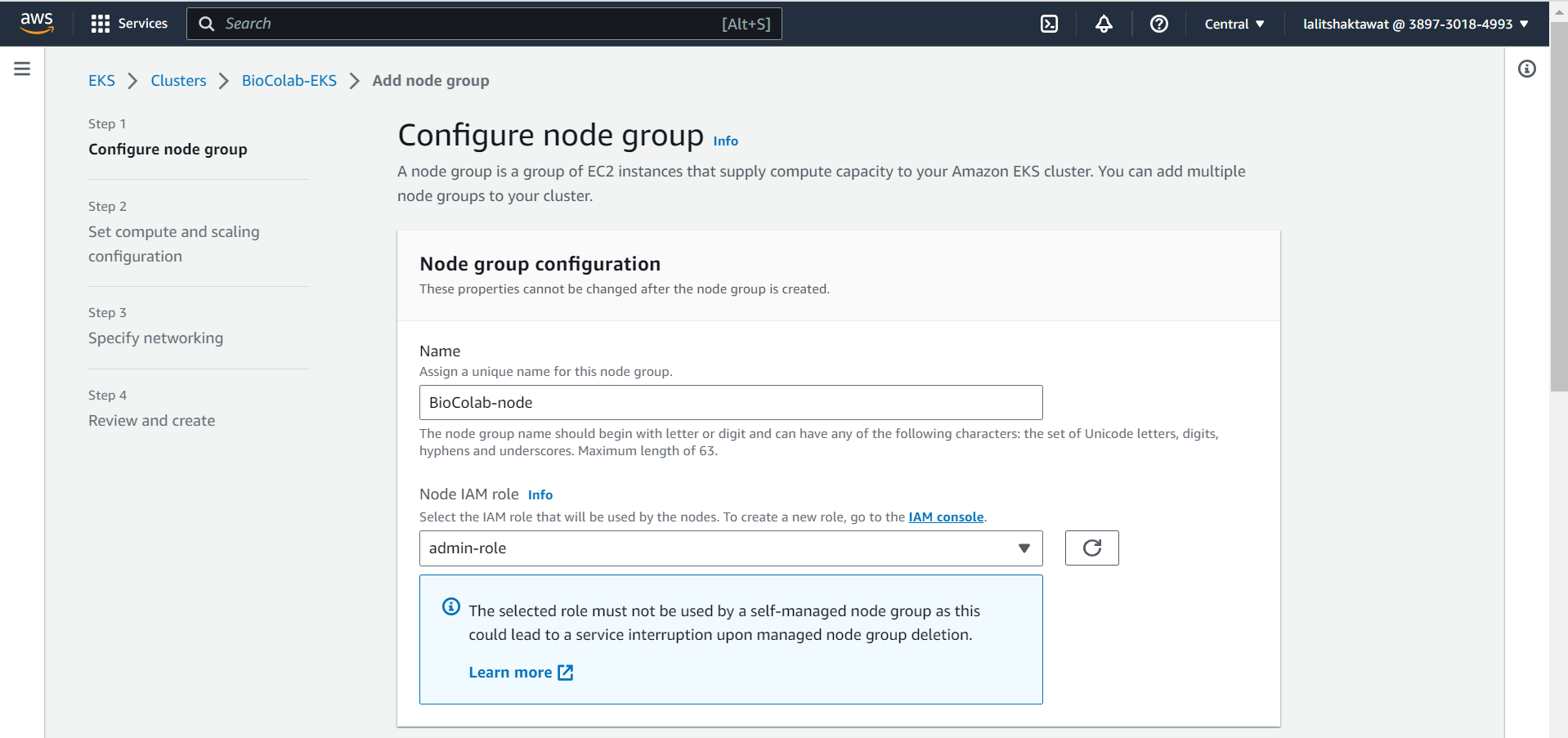
⭕ Click on Create role.
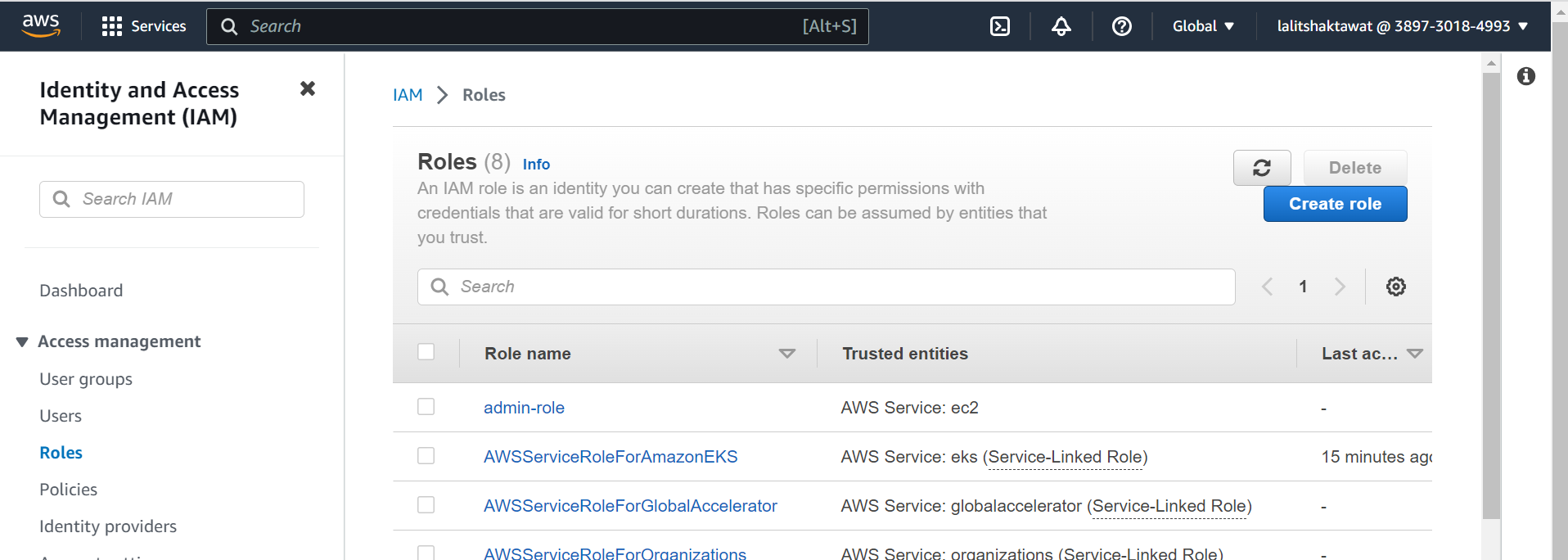
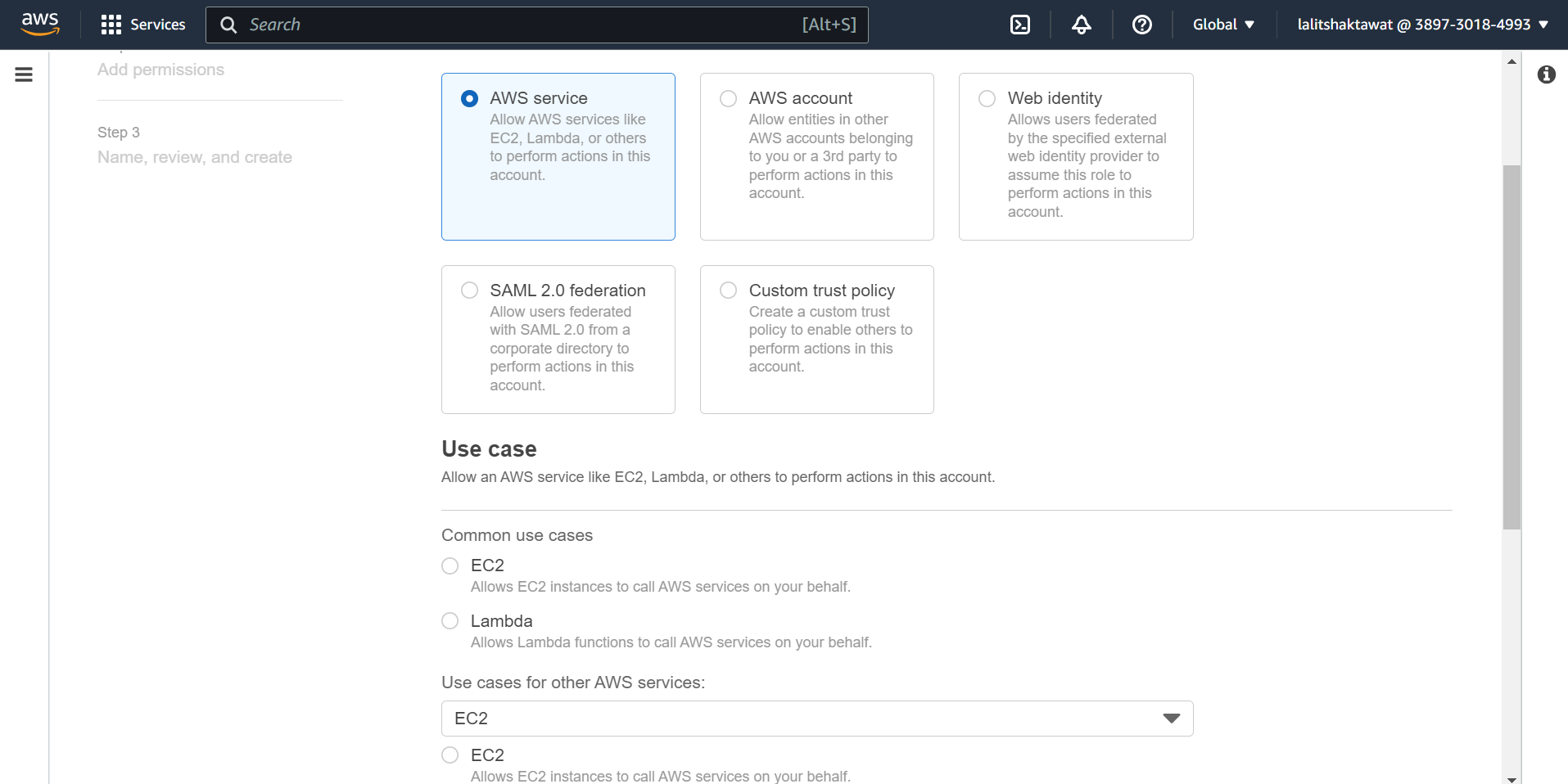
⭕ Select AWS service.
Use cases for other AWS services. EC2
Add three tick mark policy that role.
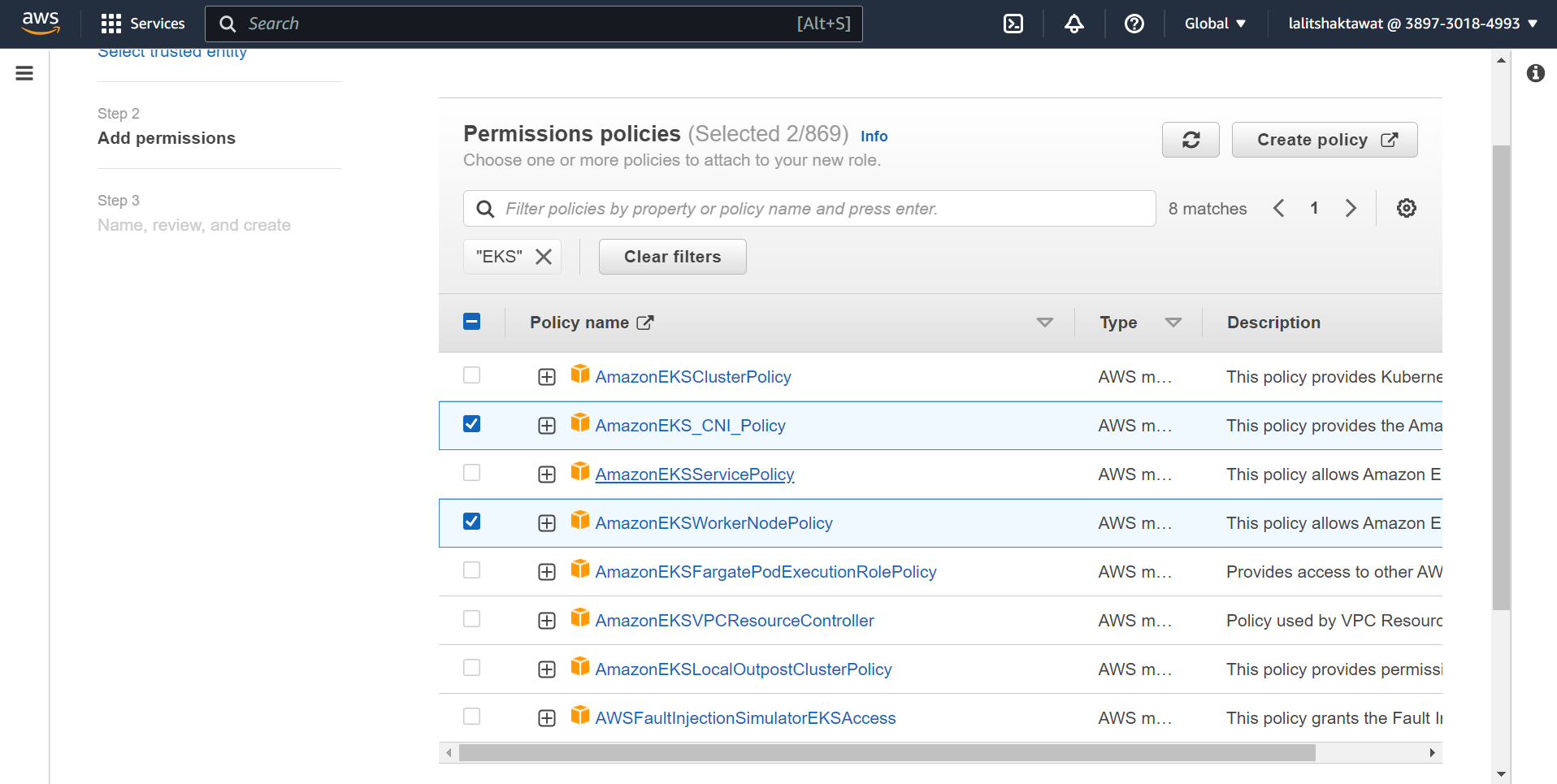
⭕ Click on Create role.
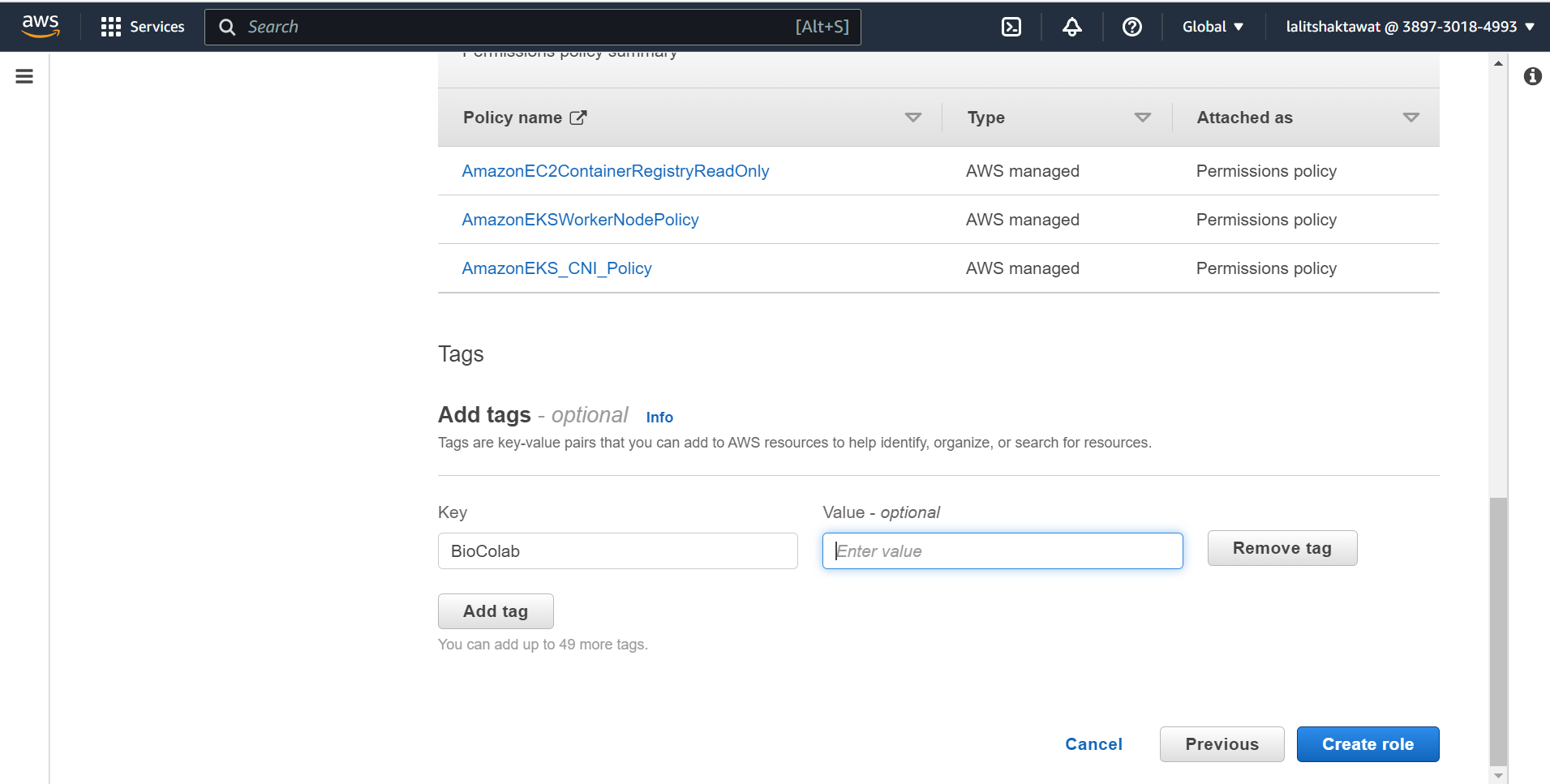
Once role is ready. Now time to assign this role to node group.
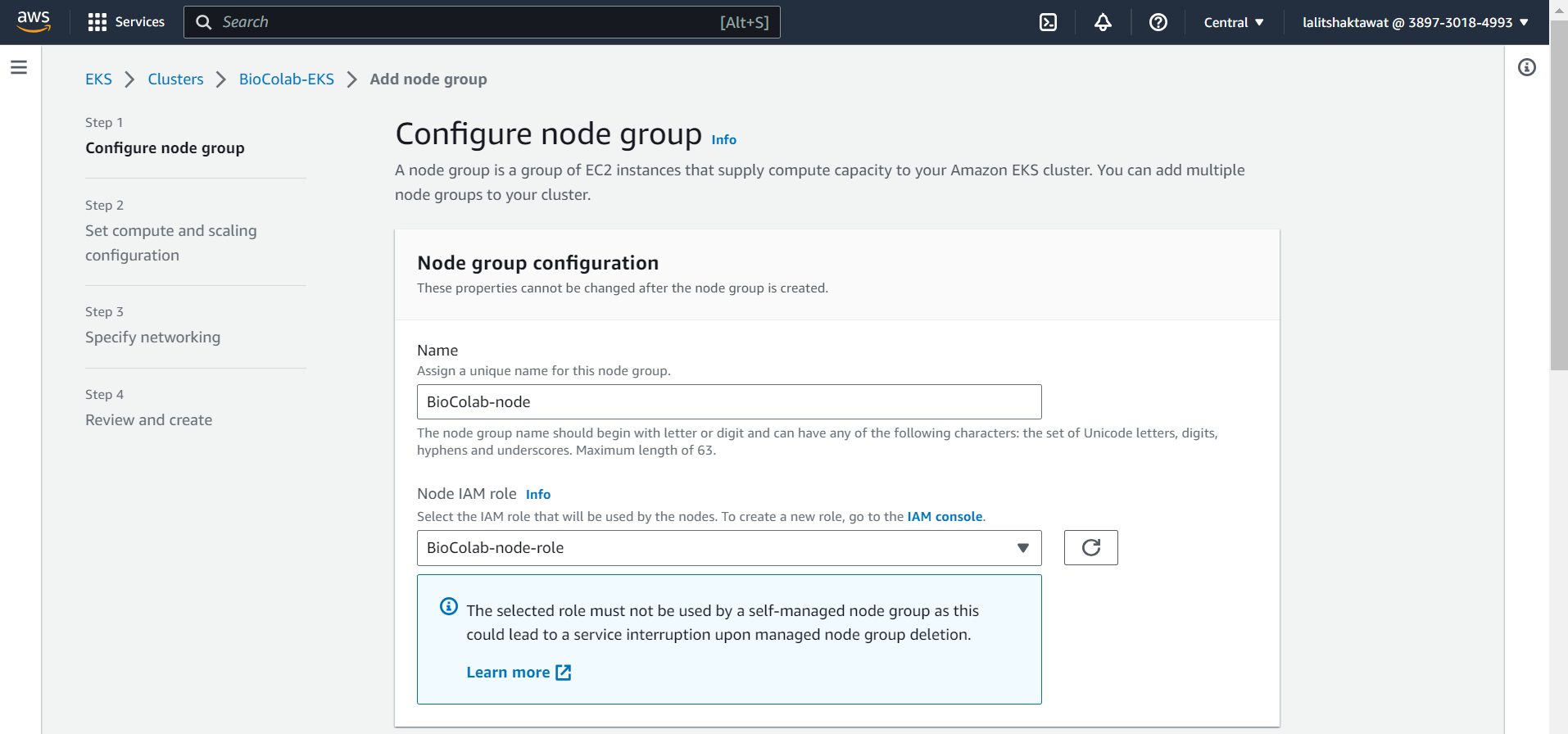
You have two option to create Ec2 instance.
1. Launch from EKS node configuration wizard.
2. Using Template
1️⃣ Launch from EKS node configuration wizard.
⭕ Click on Next.
⭕ Click on Next.
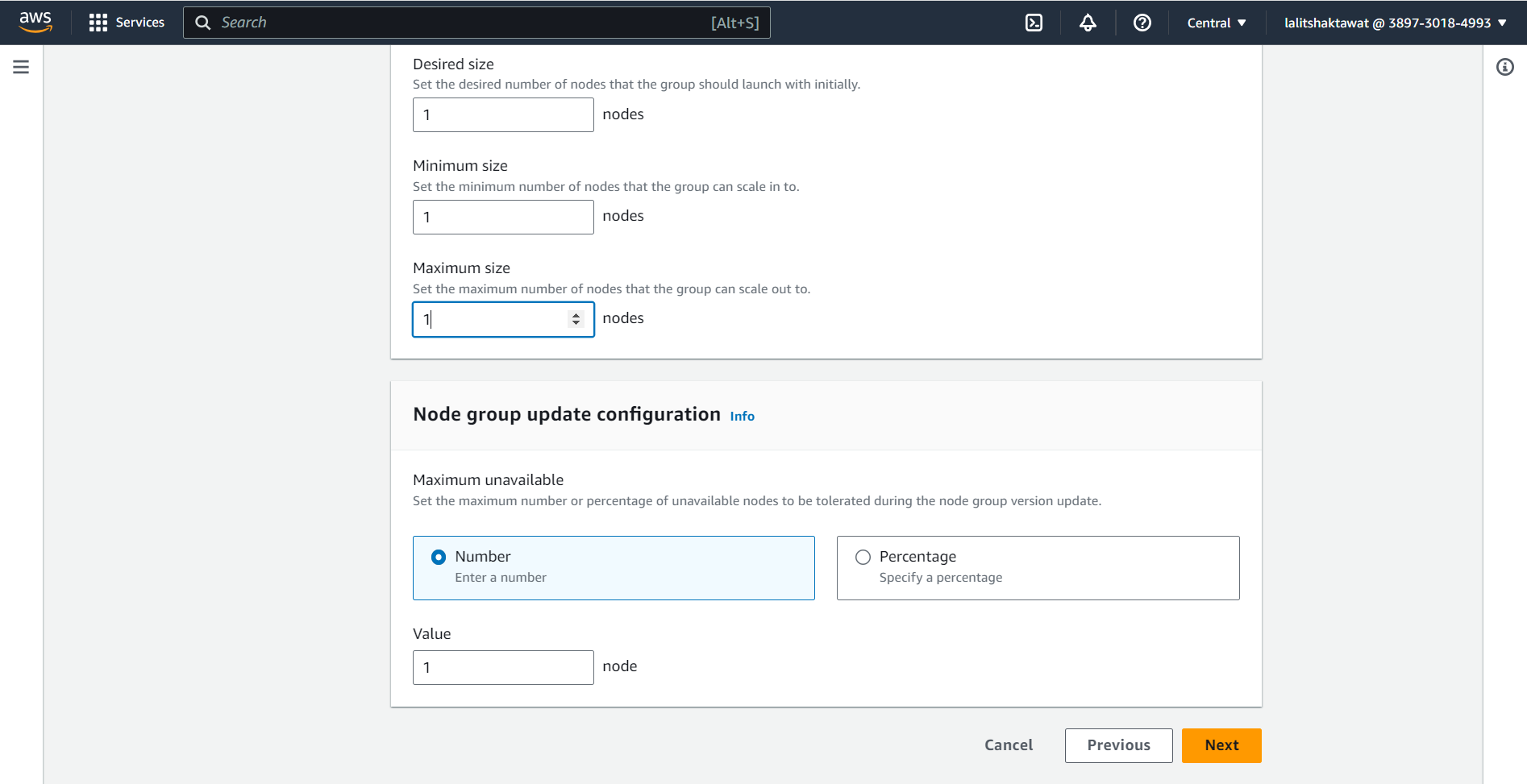
⭕ Click on Next.
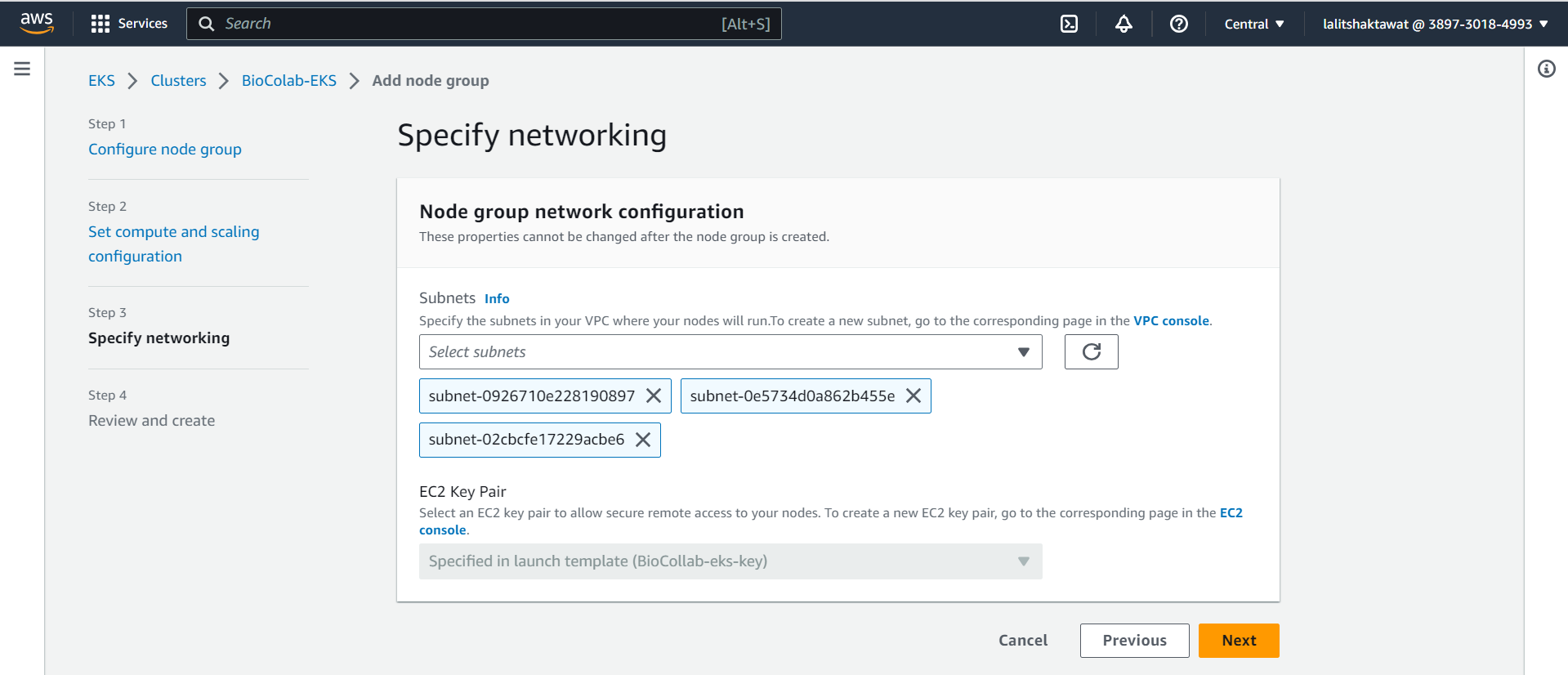
⭕ Click on Create.
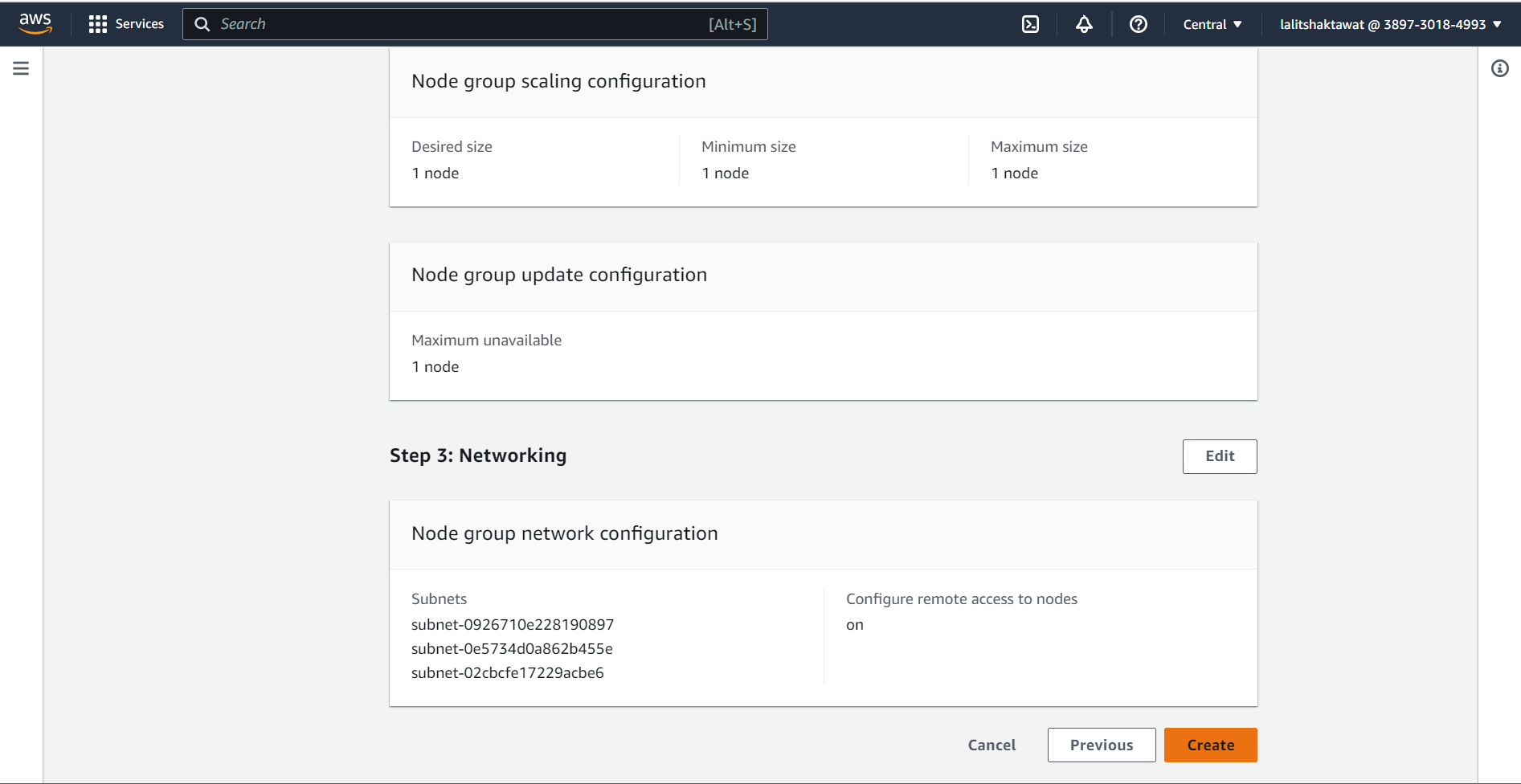
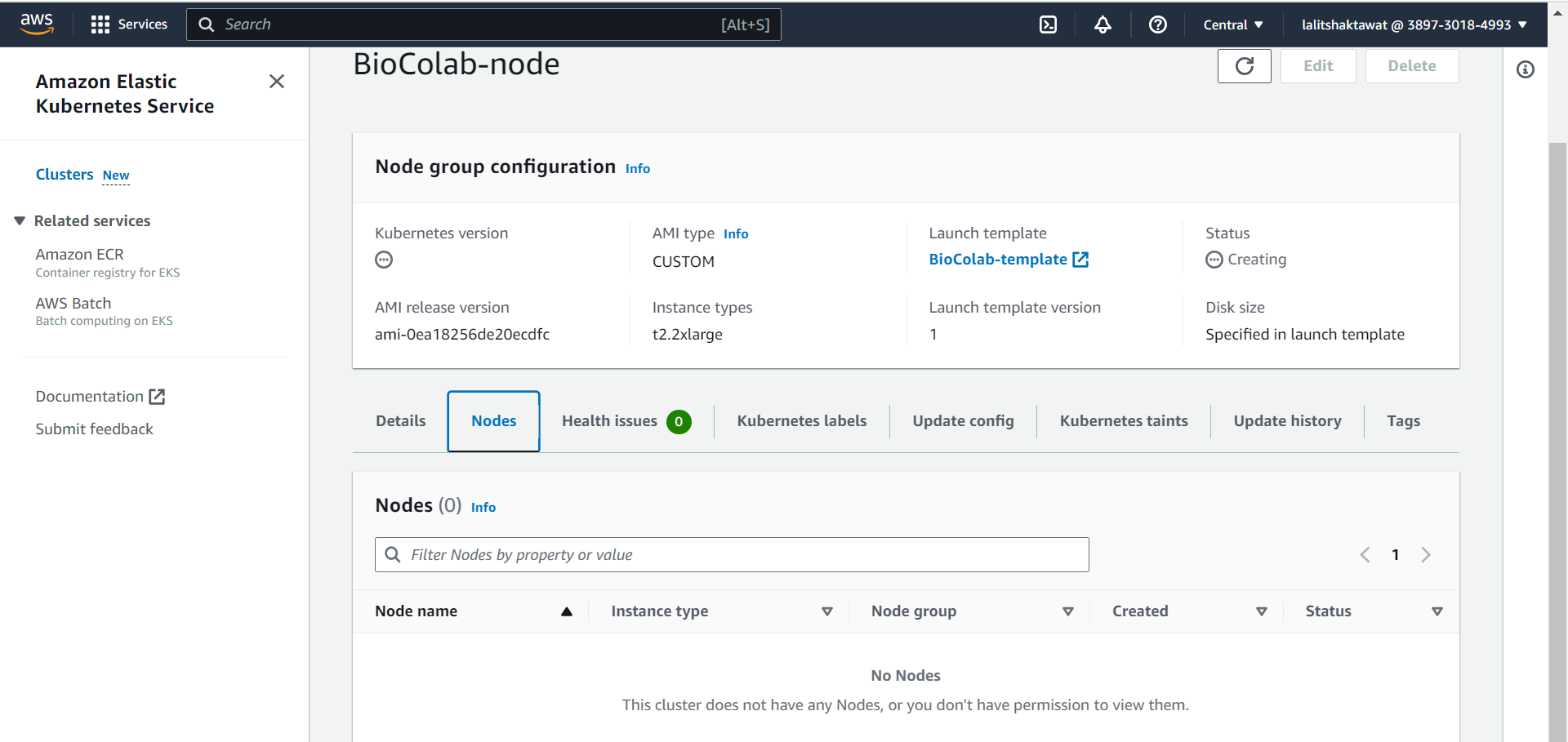
🔄 Wait for a while to add node to the Cluster.
2️⃣ Add node group using template wizard.
⭕ Click on Create Launch template.
▶️ Follow the instruction based on Template creation steps.
⭕ Once followed to provide all the value. Click on create launch template.
⭕ Once template ready. Add to the node group.
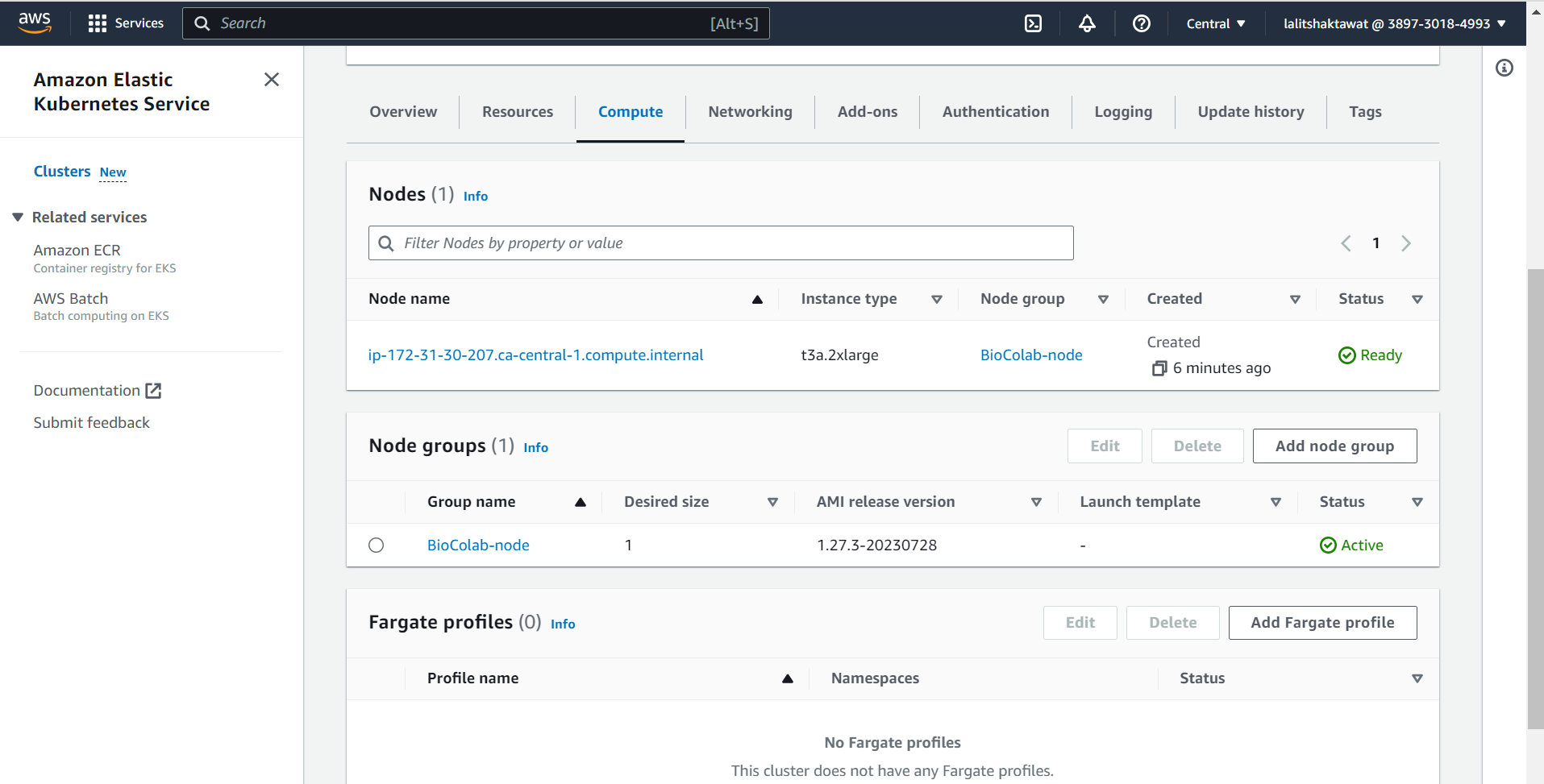
⭕ In order to use PVC creation. We must have driver installed with cluster.
⭕ Click on Add-ons tab.
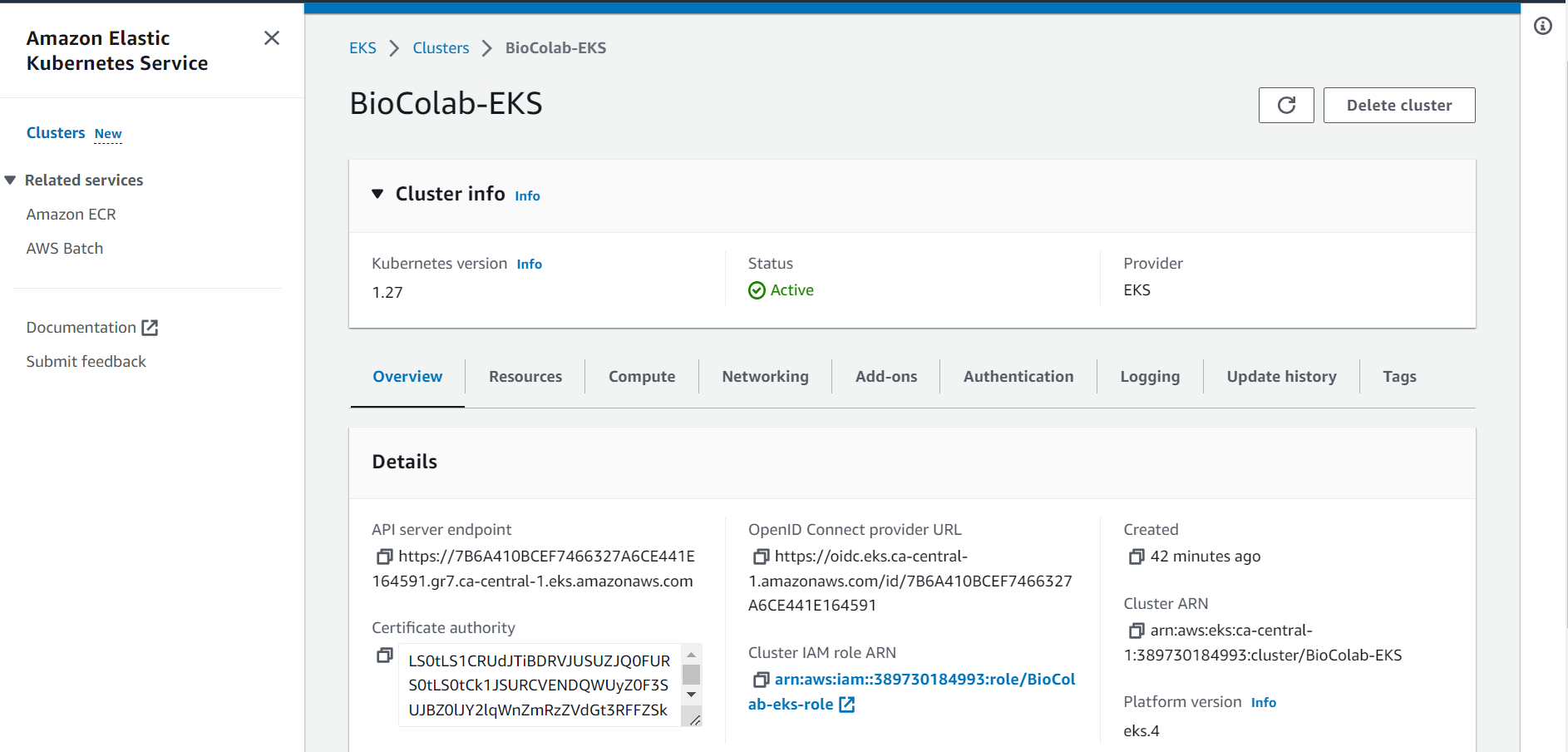
⭕ Click on Get more Add-ons.
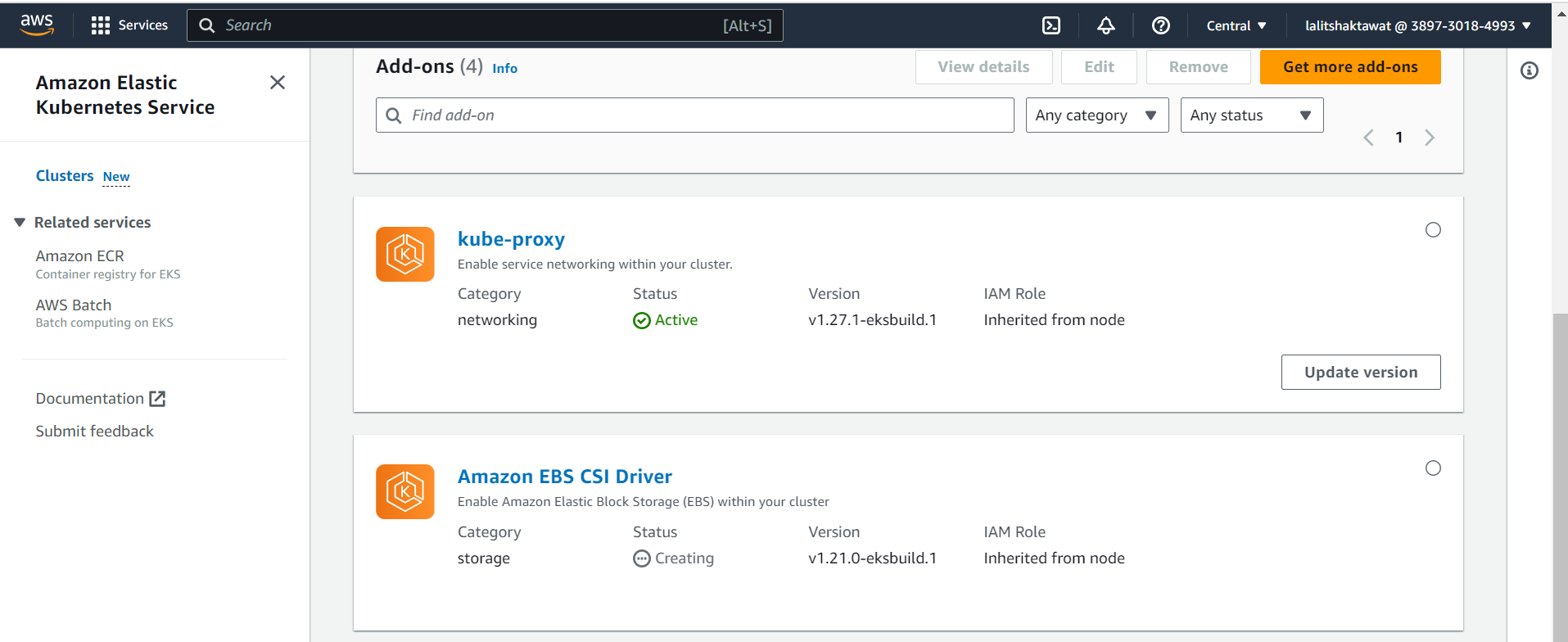
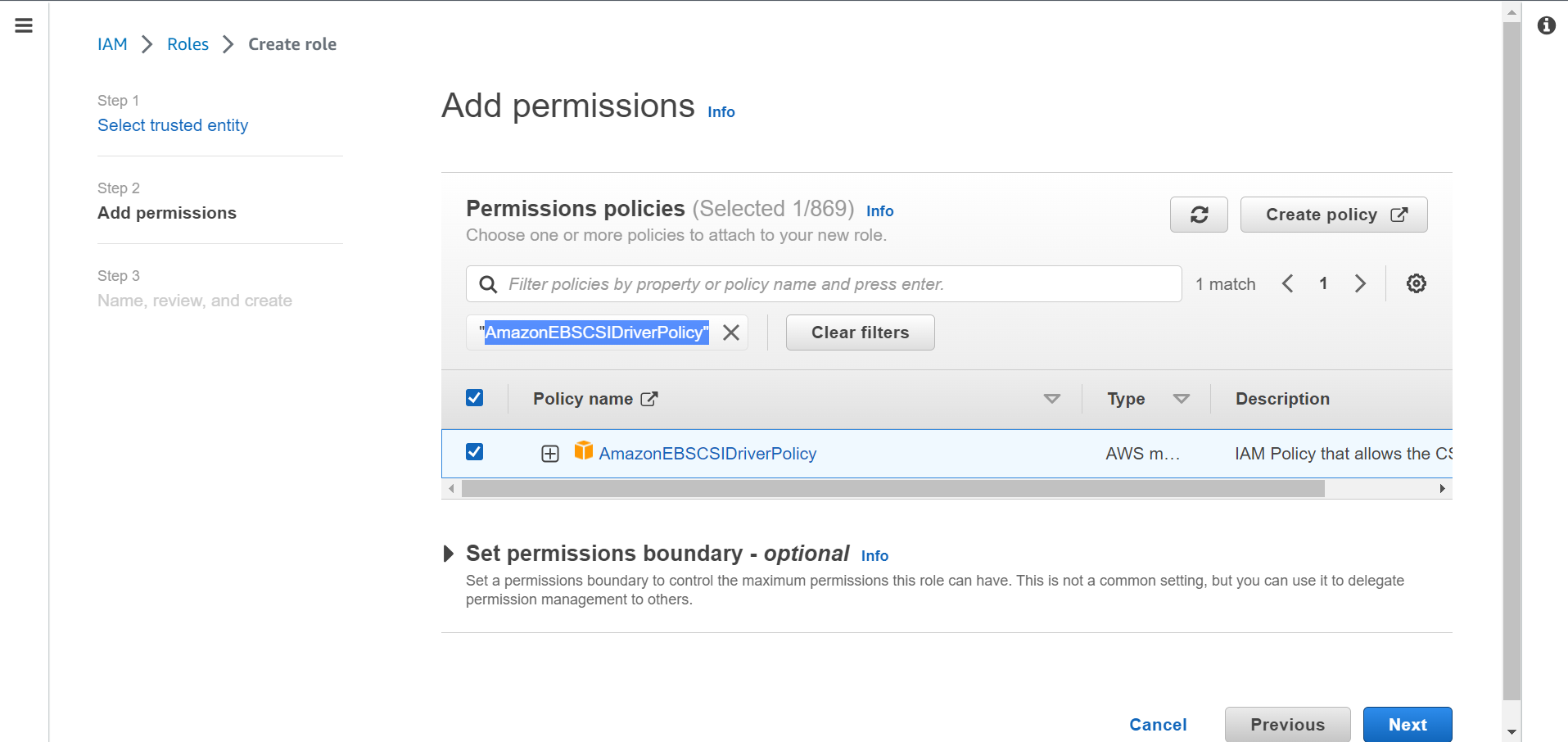
⭕ Select Amazon EBS CSI Driver.
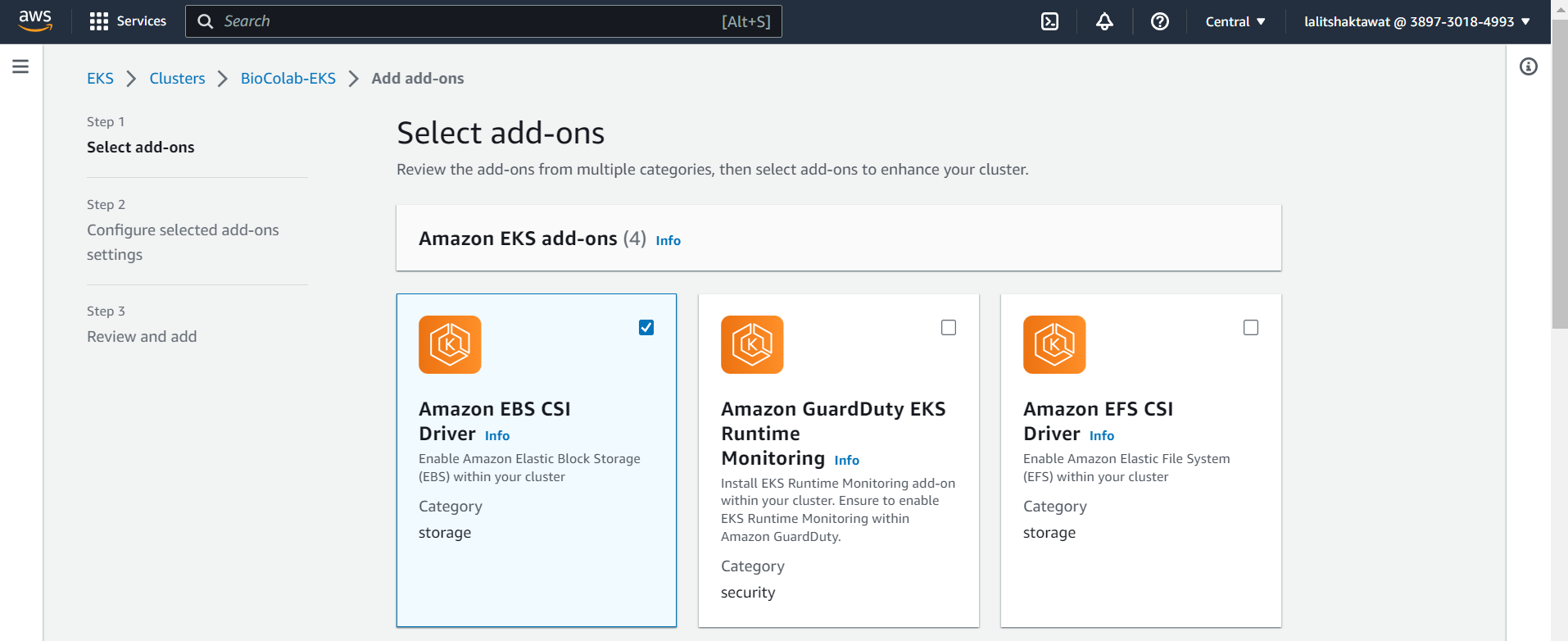
⭕ Click on next.
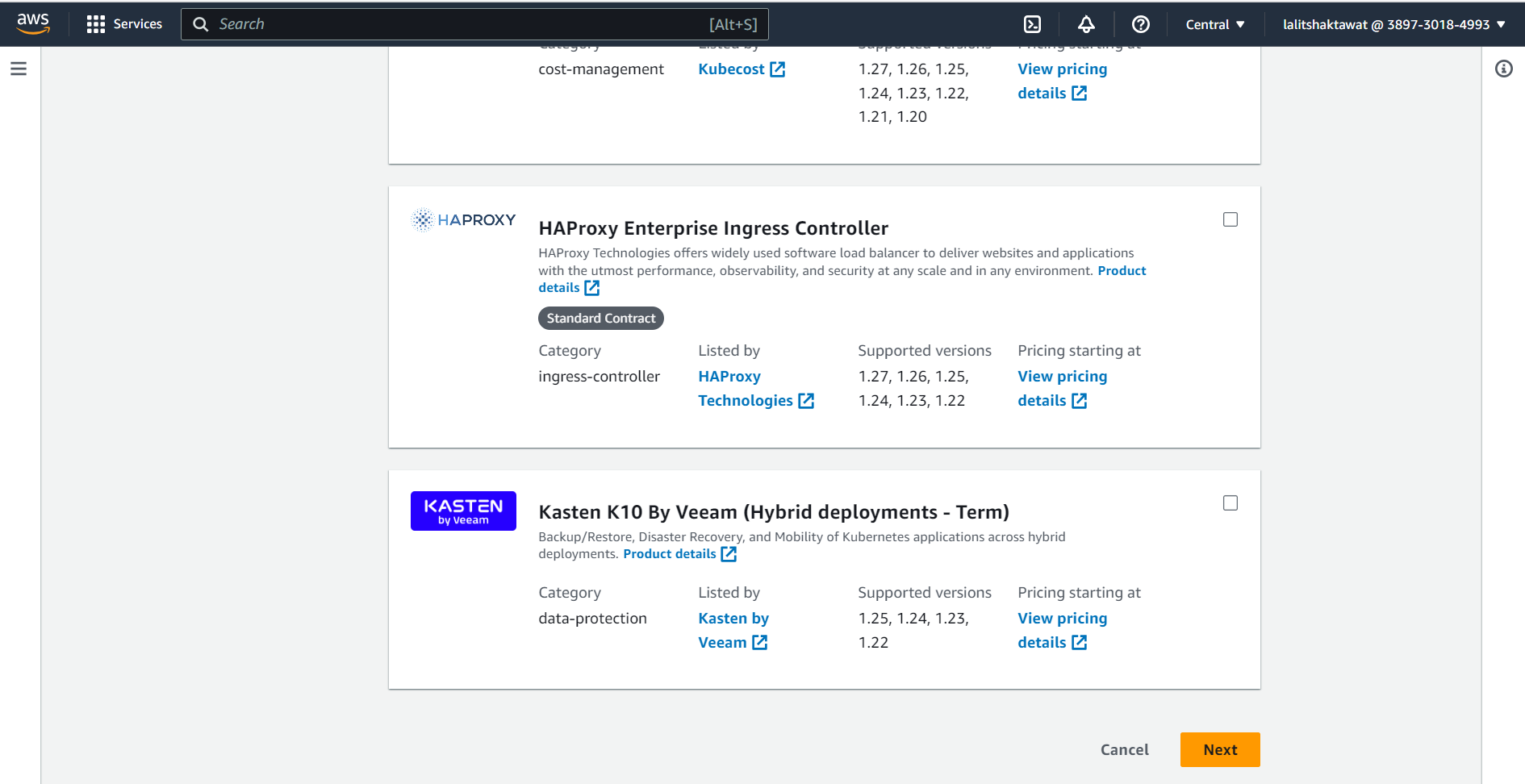
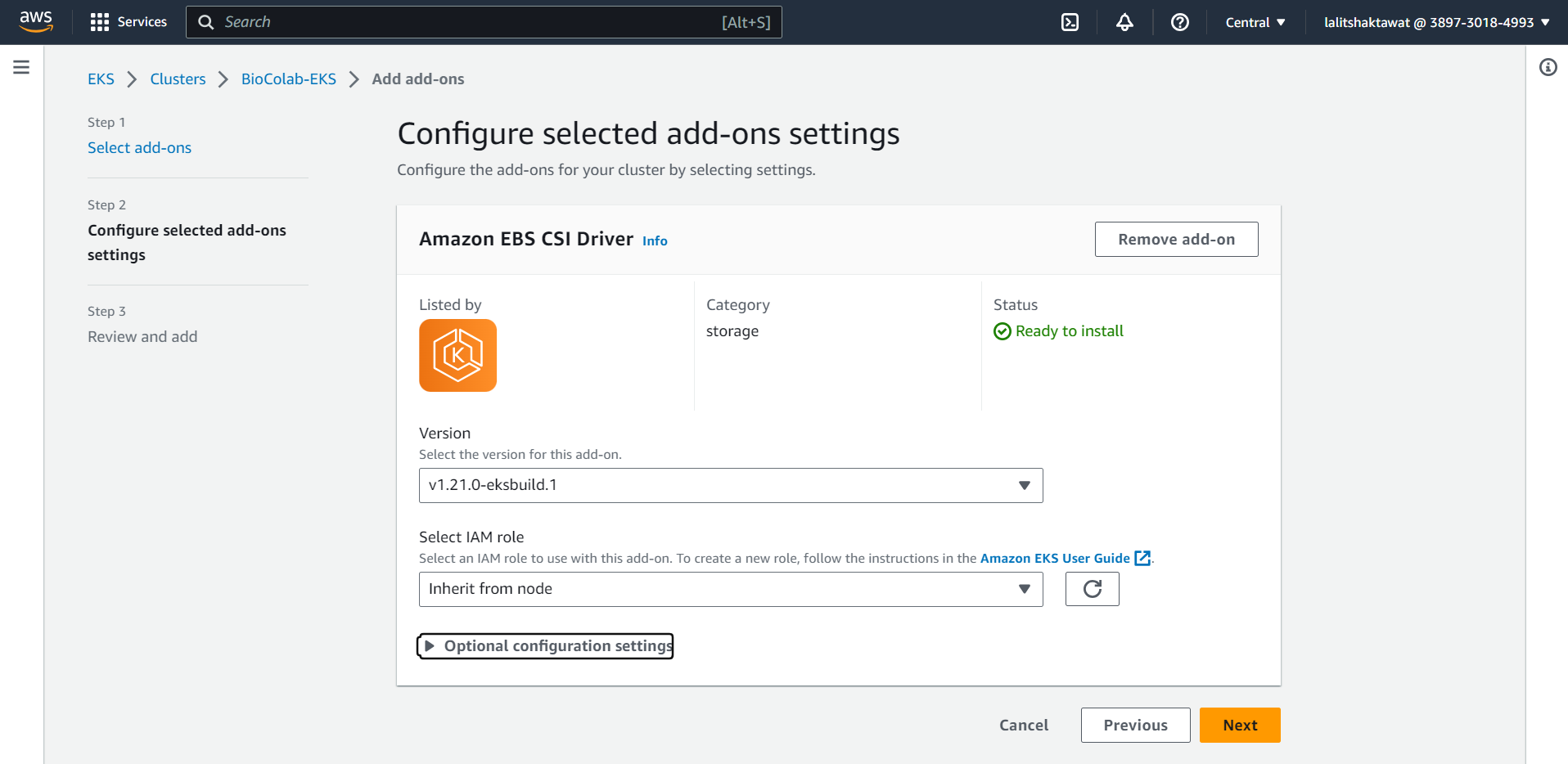
⭕ Click on Create.
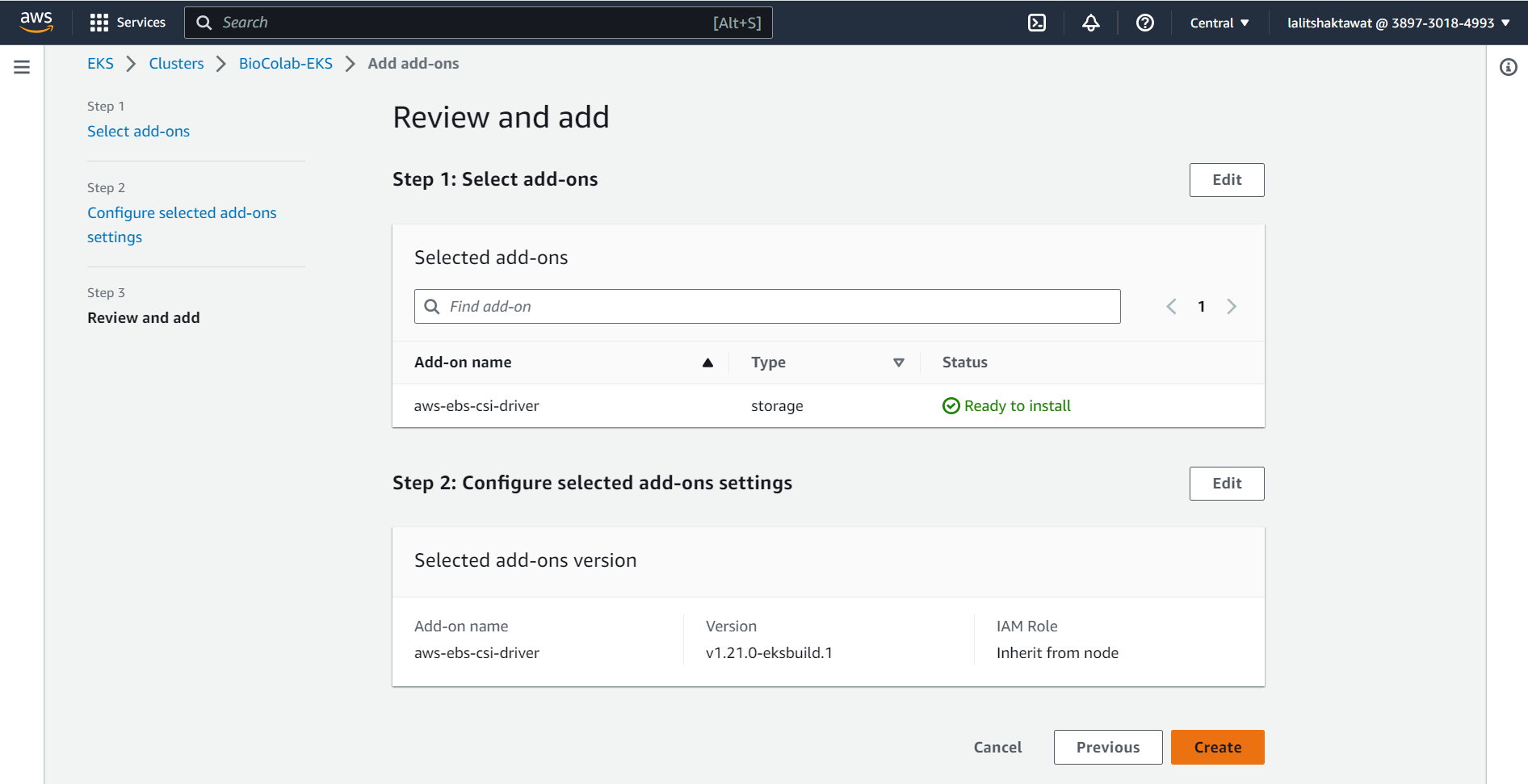
✴️ We need to create IAM OIDC provider for your cluster.
🔗 OIDC
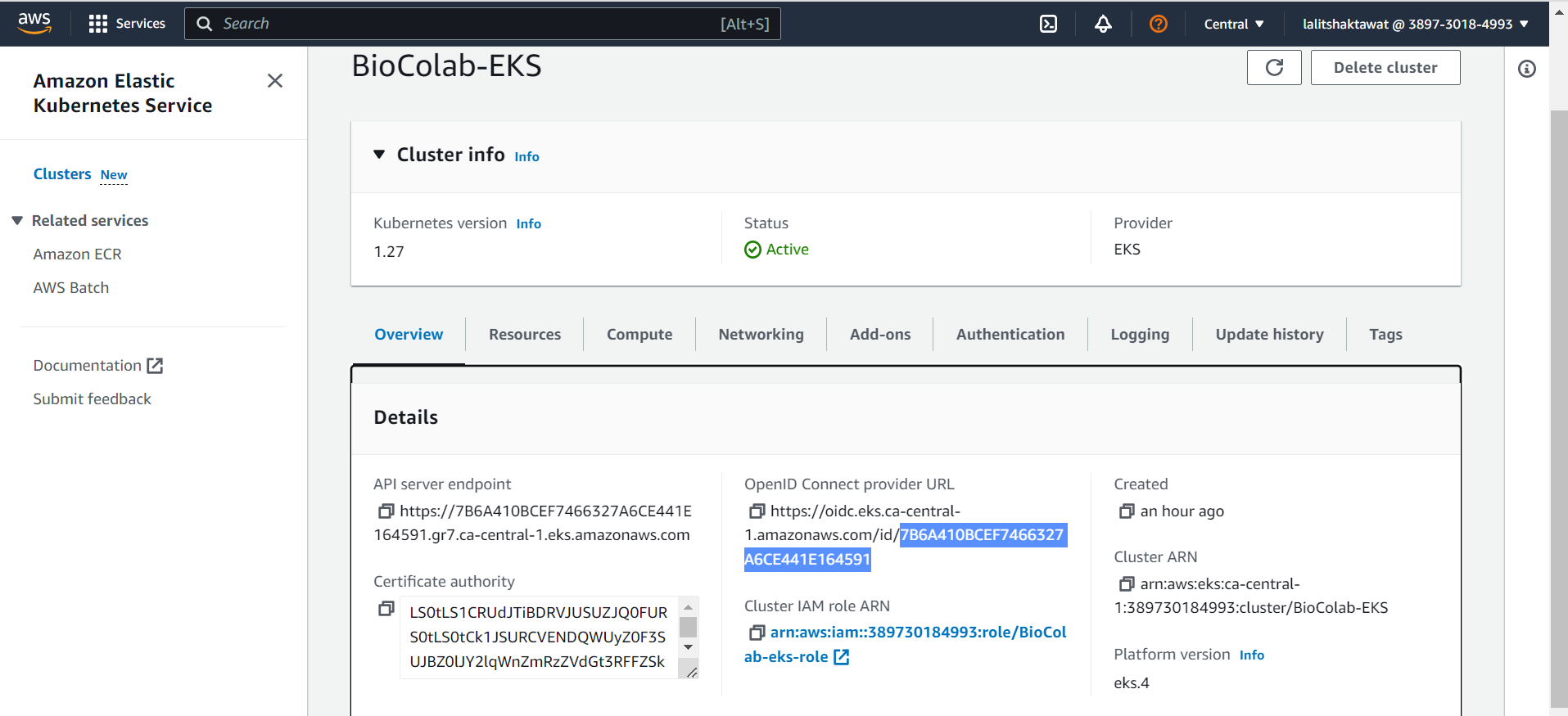
⭕ Copy the OpenID Connect provider URL.
➡️ Search IAM and click on role
➡️ Select Identity provider on Access management.
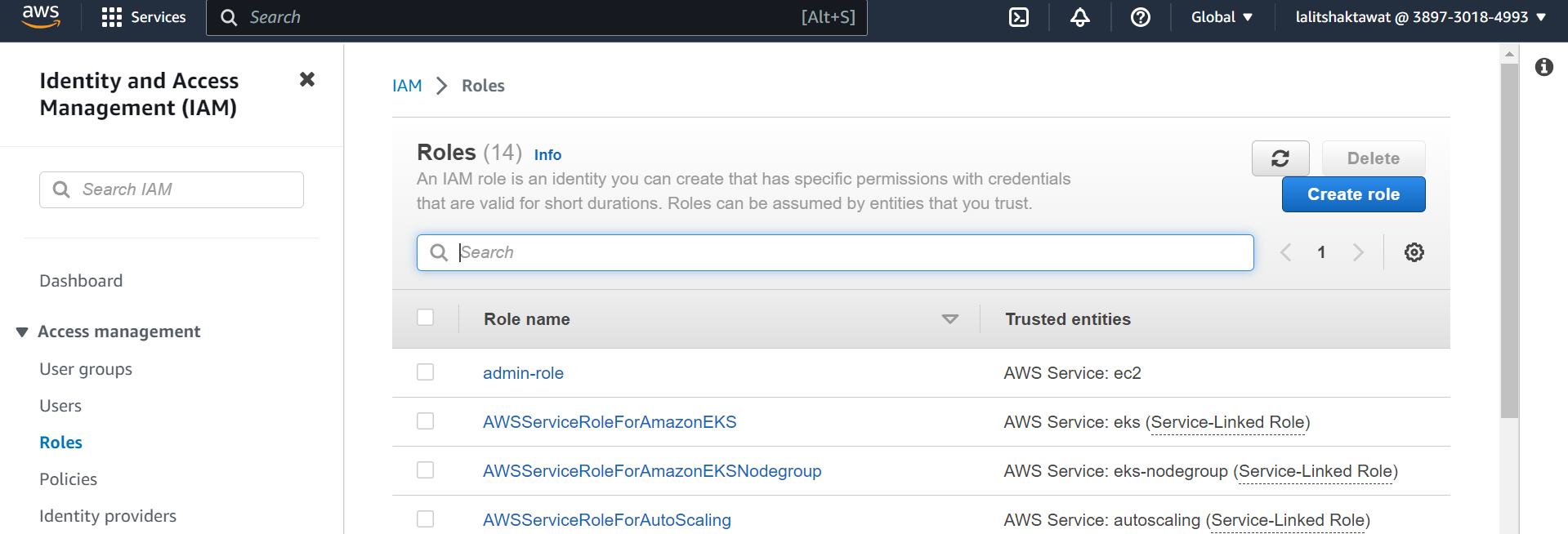
➡️ Click on Add provider.

➡️ Click on OpenID Connect.
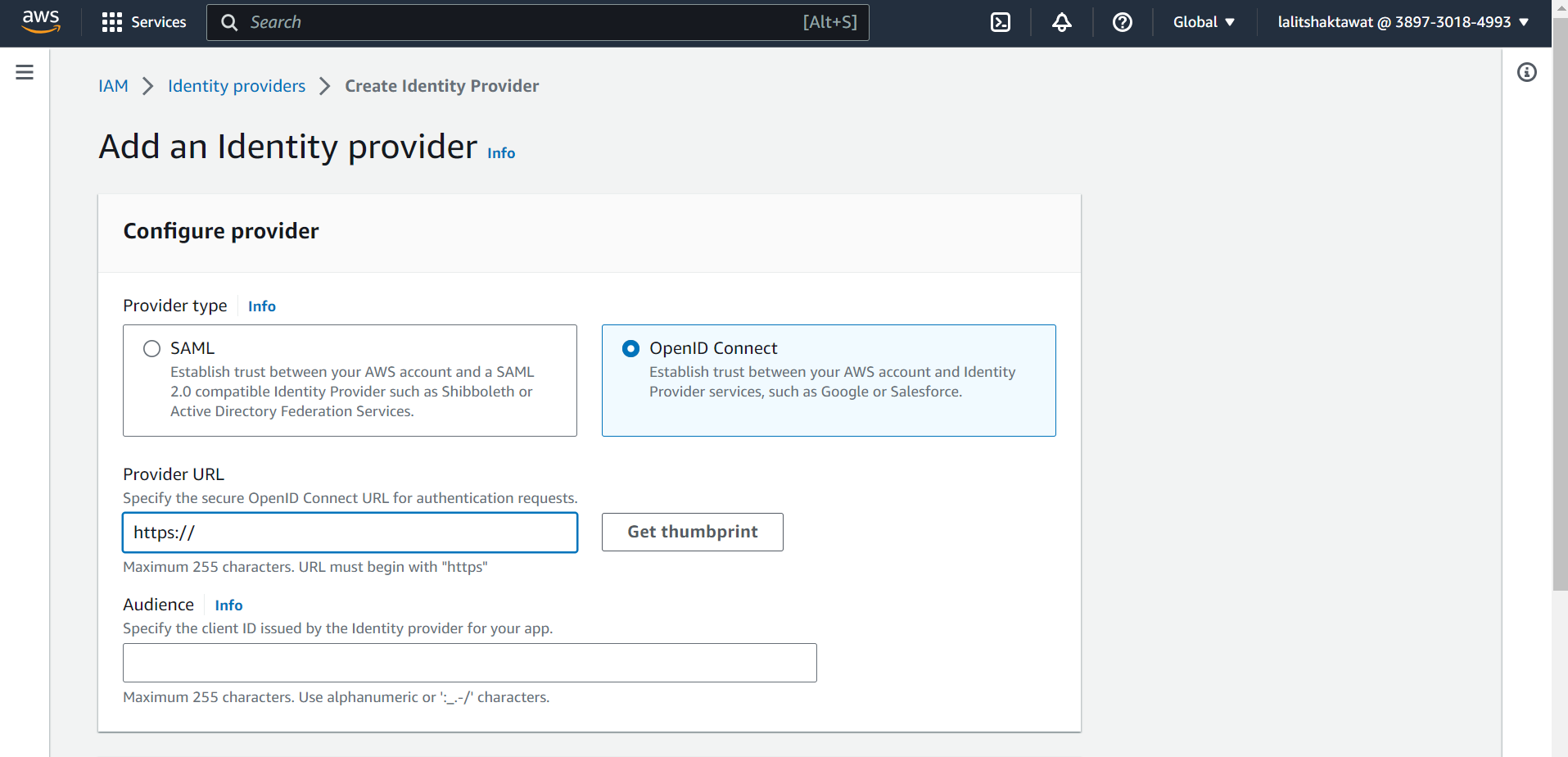
➡️ Paste Provider URL tht we copied earlier from Cluster.
➡️ Click on Get thumbprint.
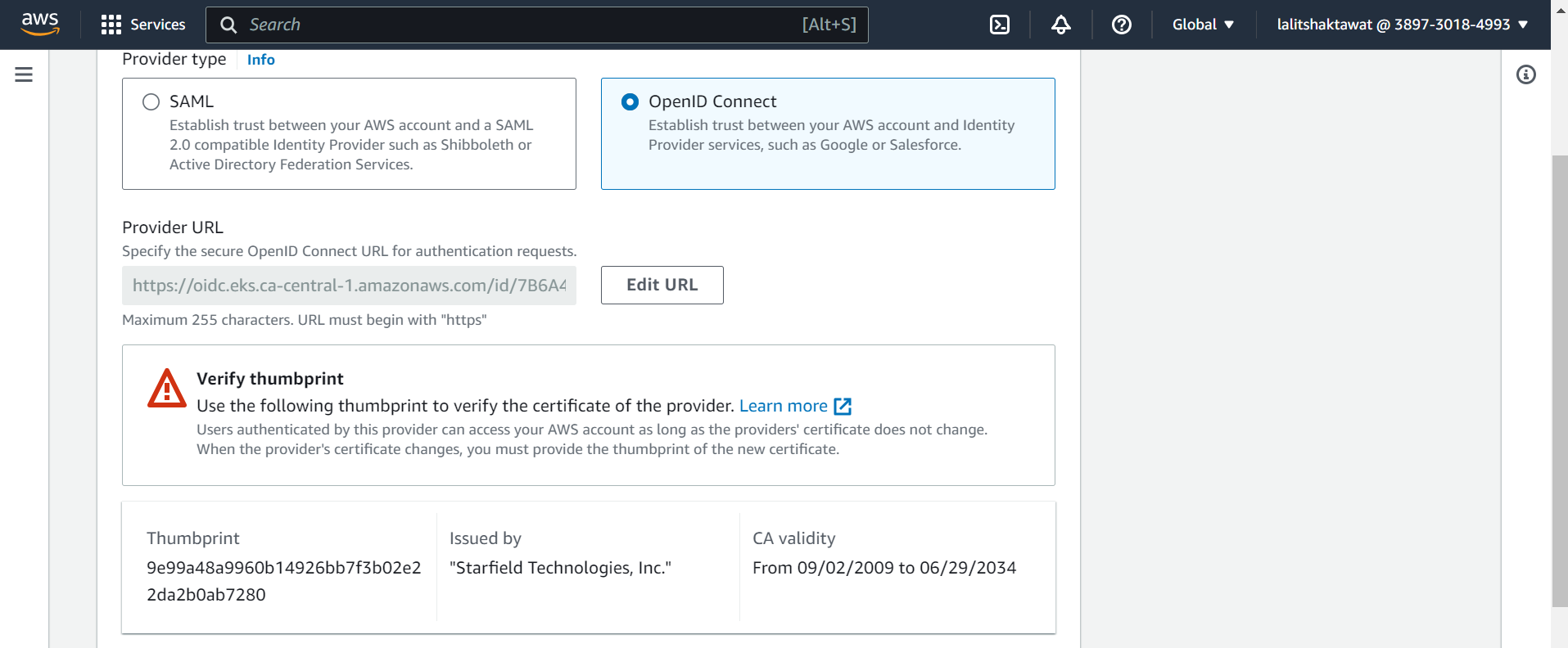
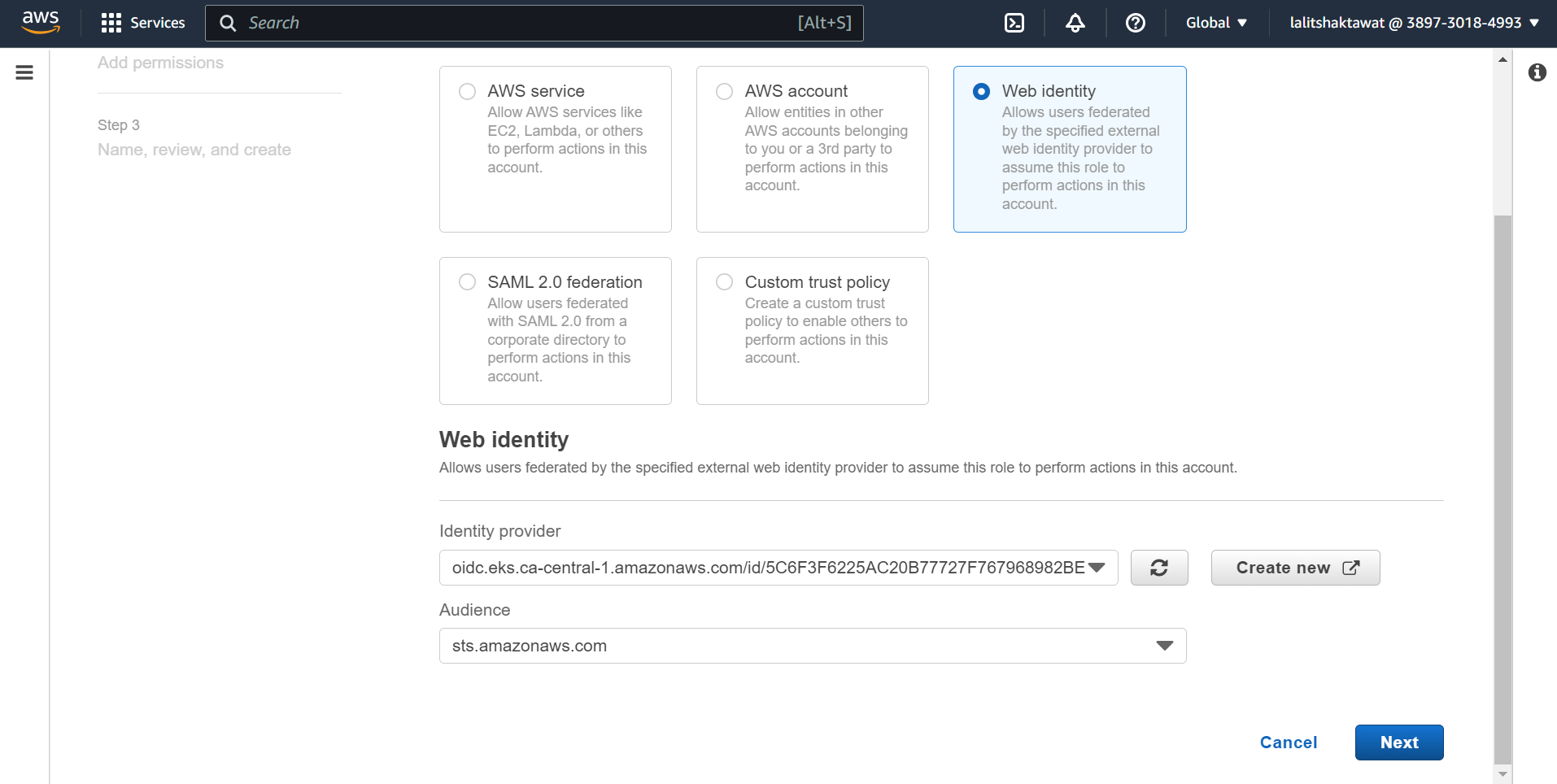
➡️ Audience, enter sts.amazonaws.com and choose Add provider.
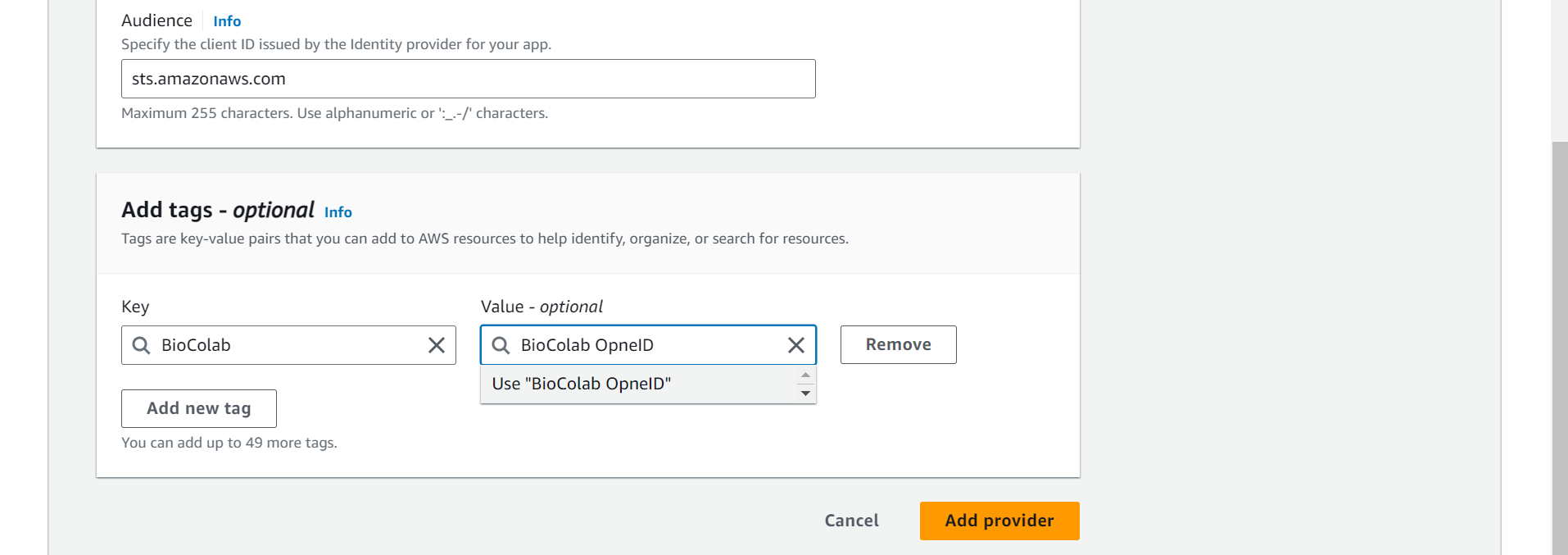
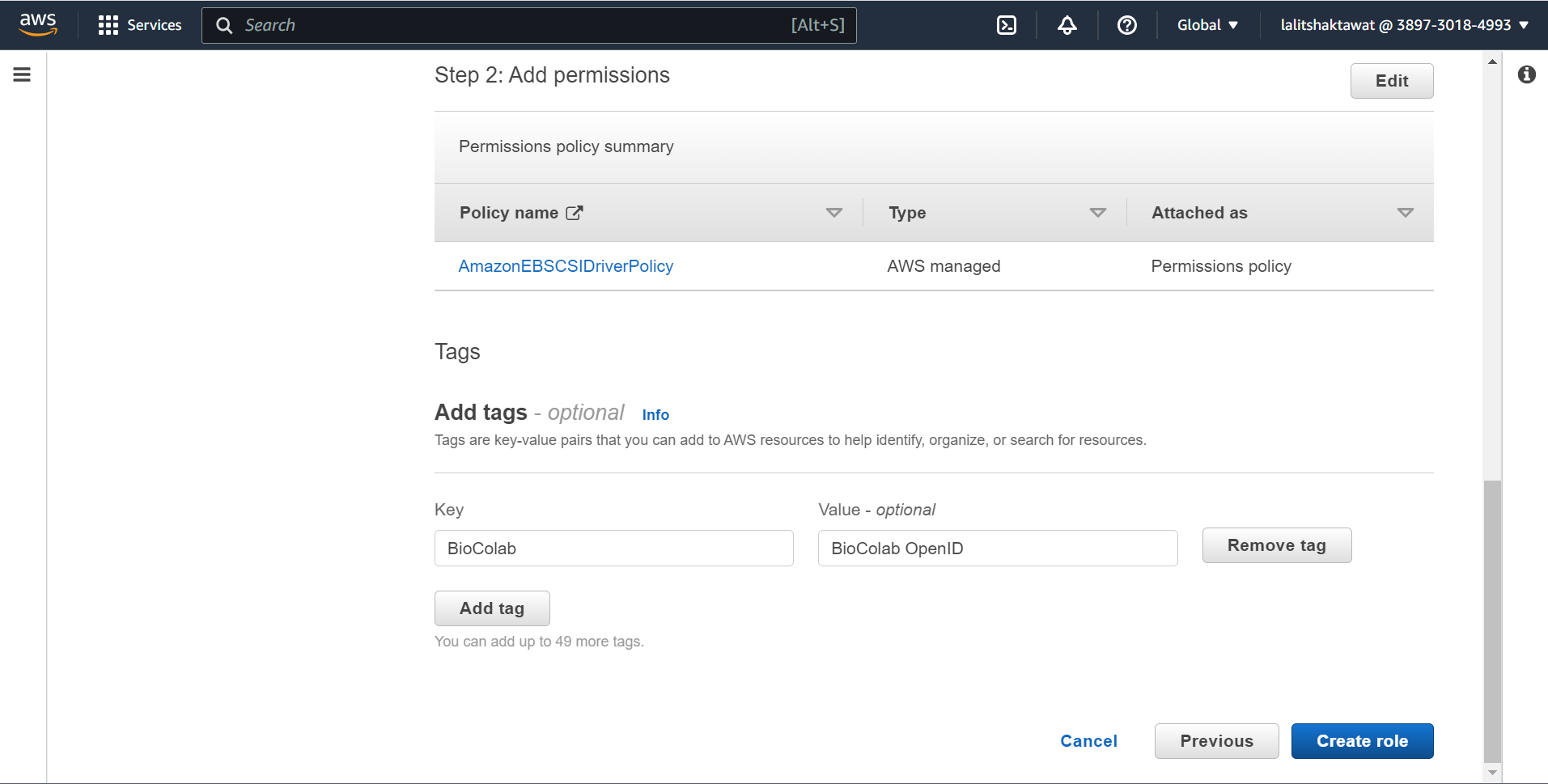
⭕ Create Amazon EBS CSI driver IAM role.
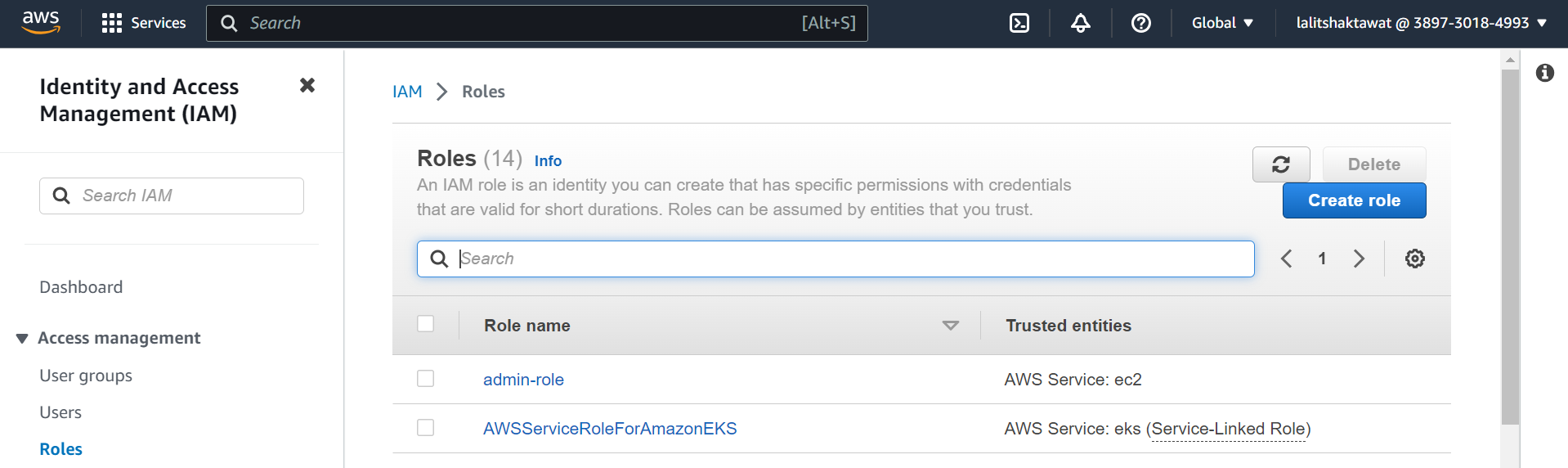
IAM --> Roles --> Create role
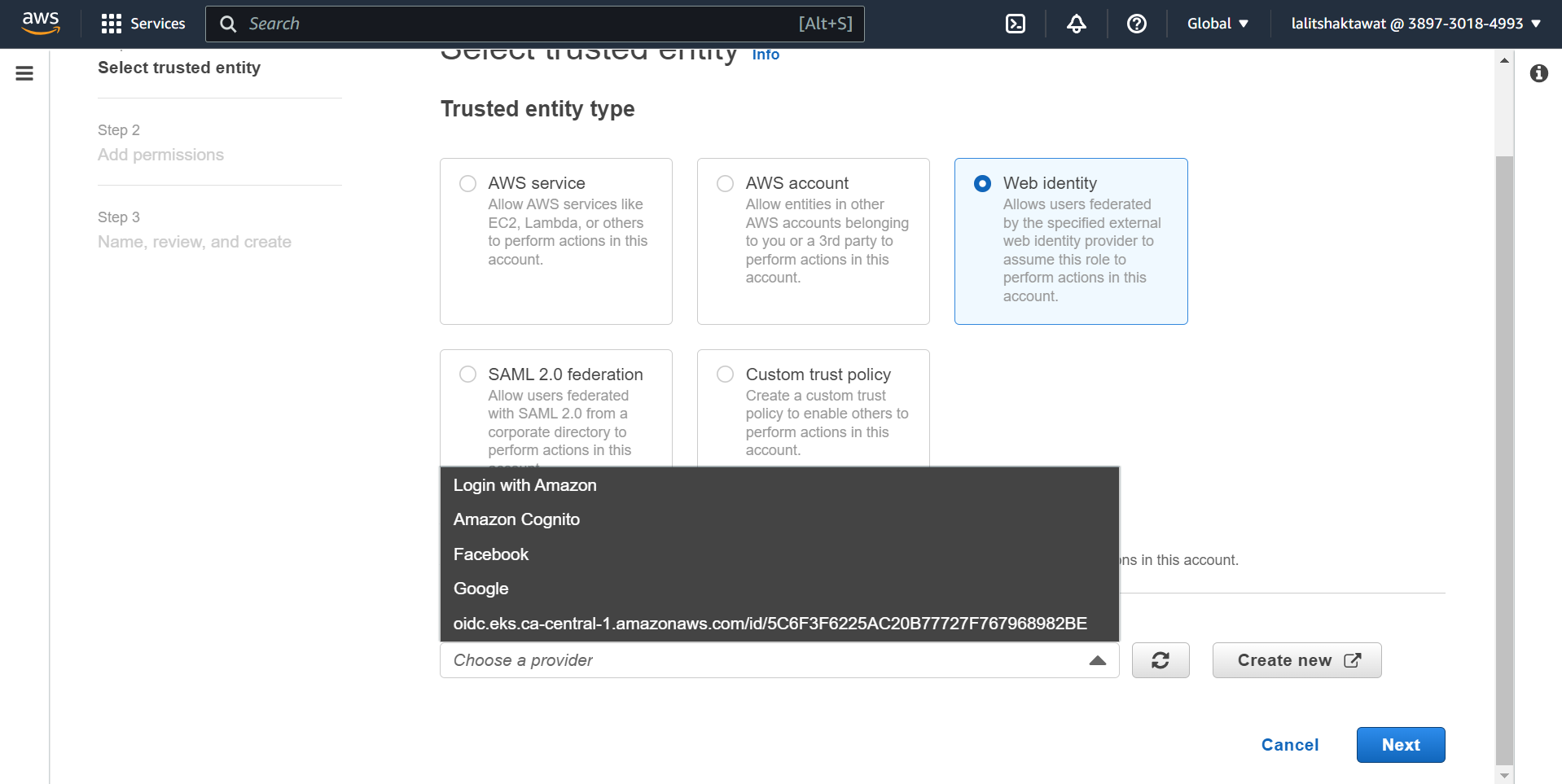
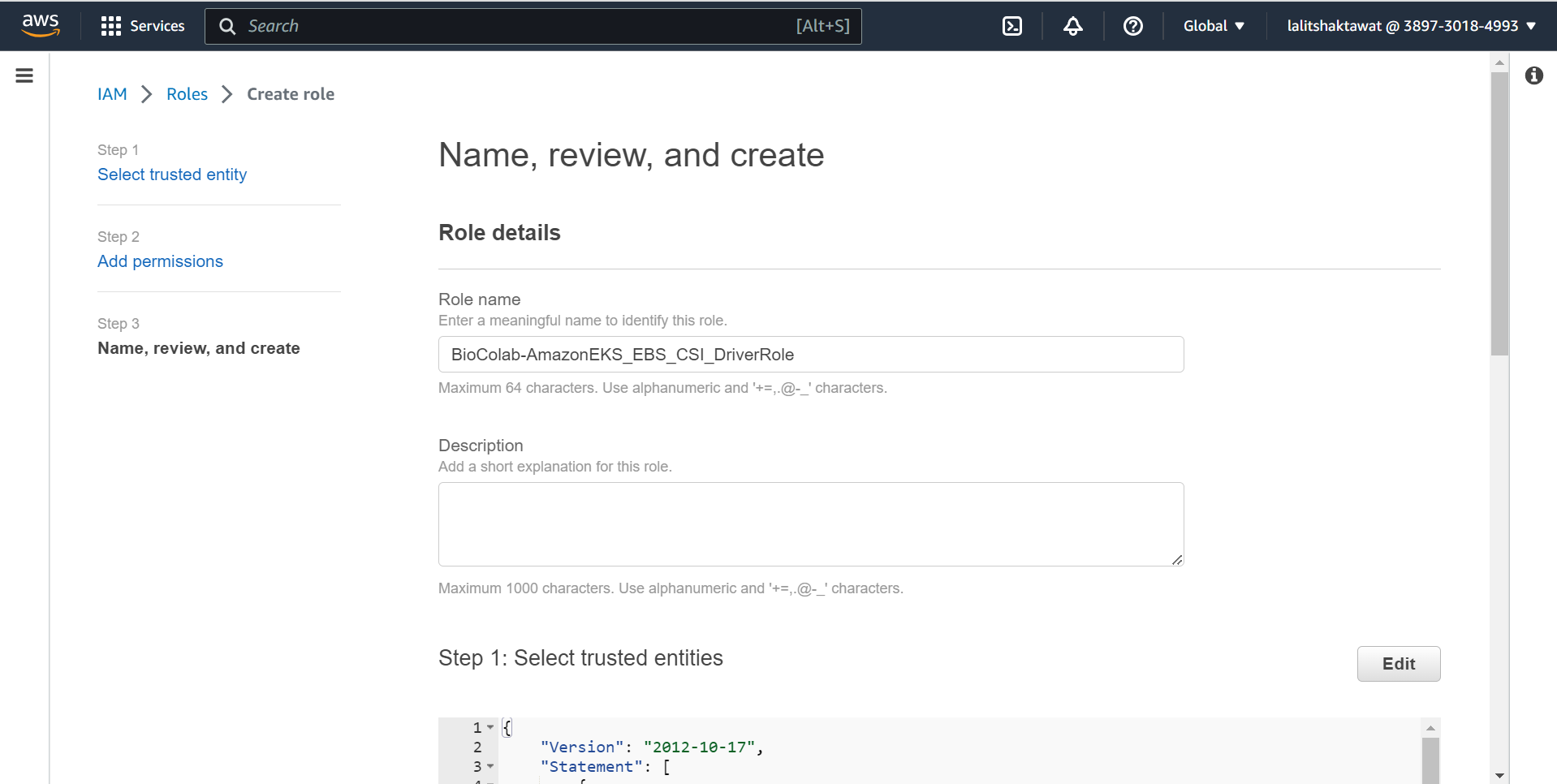
➡️ Edit trust relationship policy.
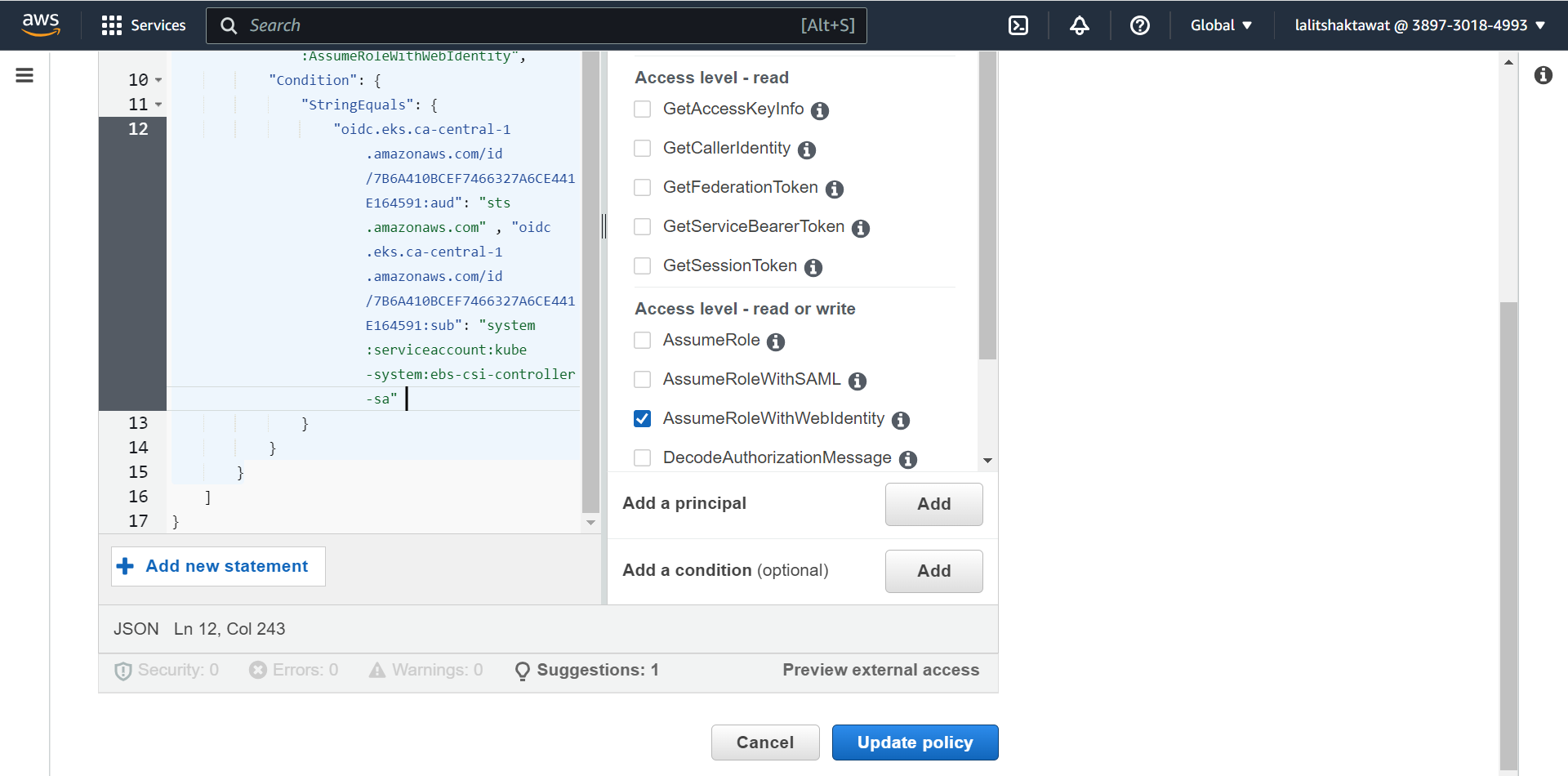
➡️ Click on Edit trust policy. Add the following line after comma
🔔 Change the value based on your OpenID
"oidc.eks.ca-central-1.amazonaws.com/id/7B6A410BCEF7466327A6CE441E164591:sub": "system:serviceaccount:kube-system:ebs-csi-controller-sa"
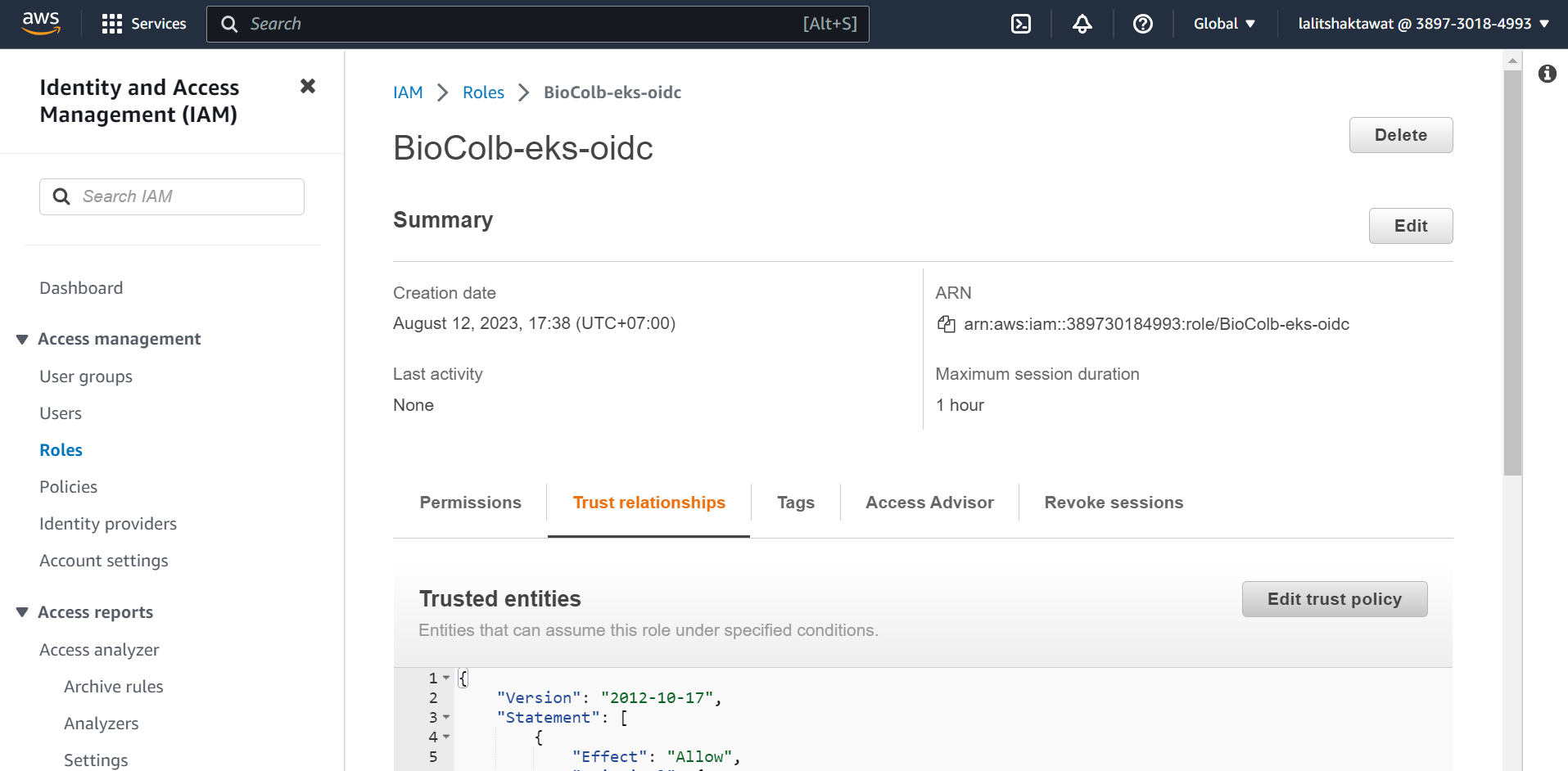
➡️ Click on Update policy.
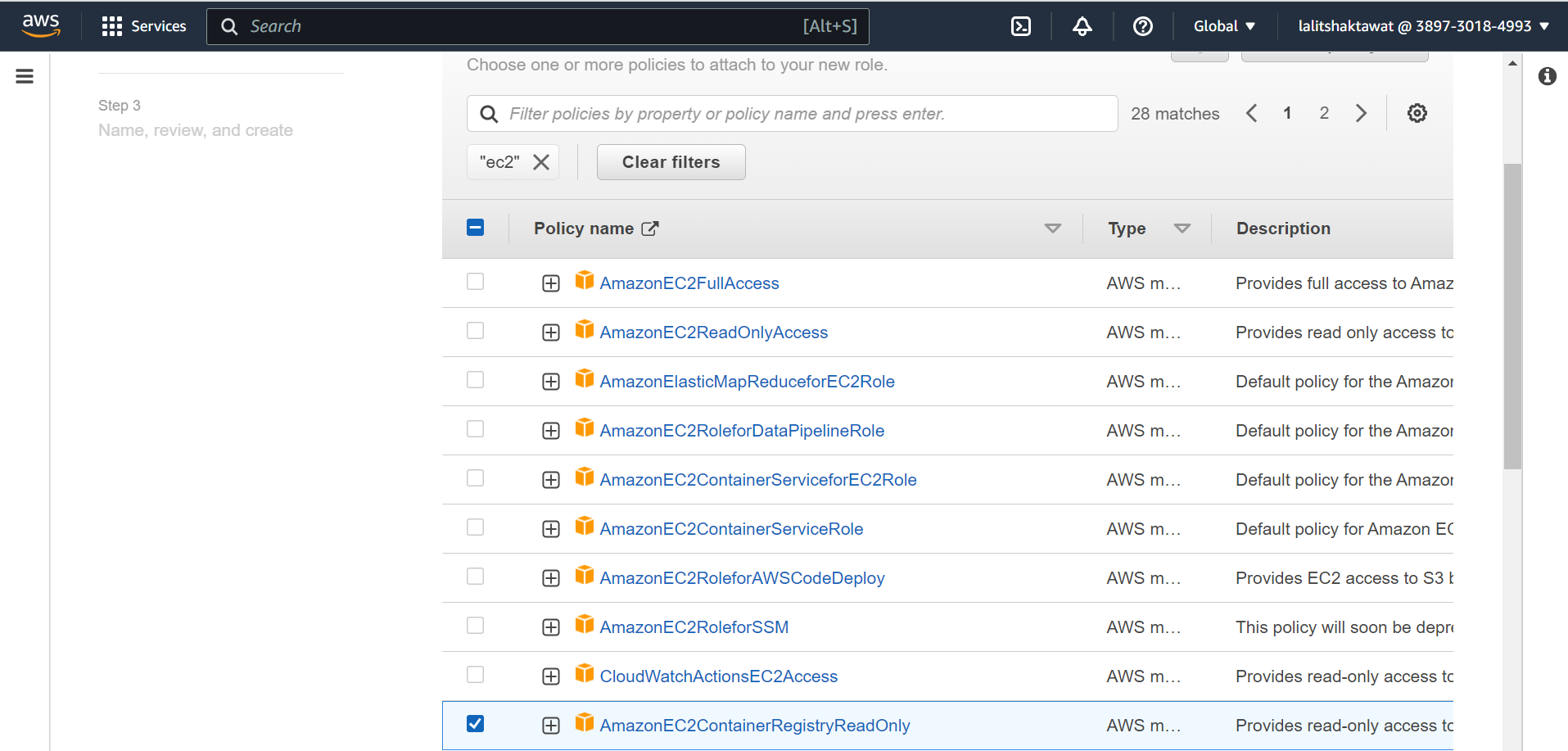
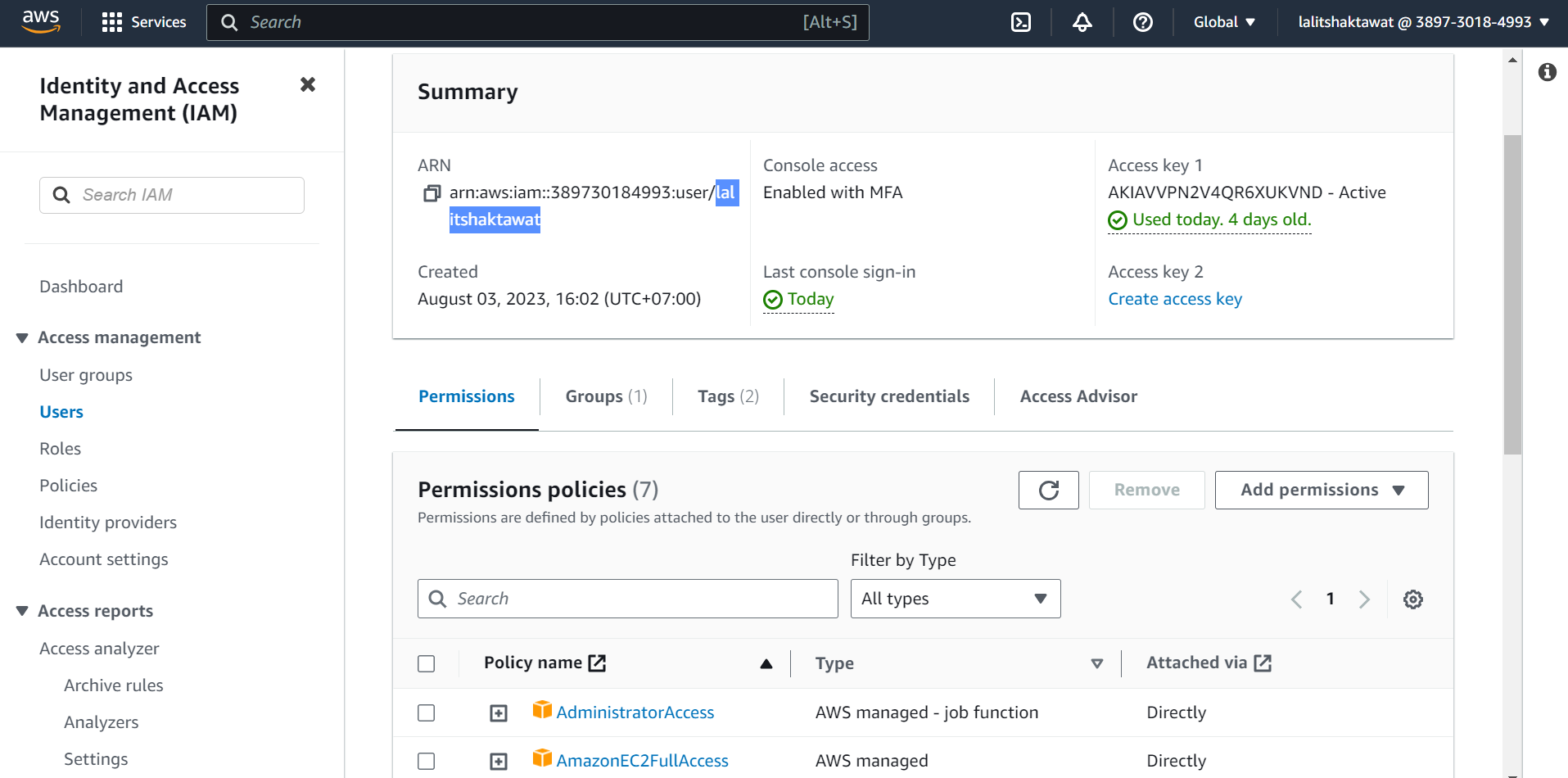
🔔 Make sure user should have permissions “AmazonEC2FullAccess”.
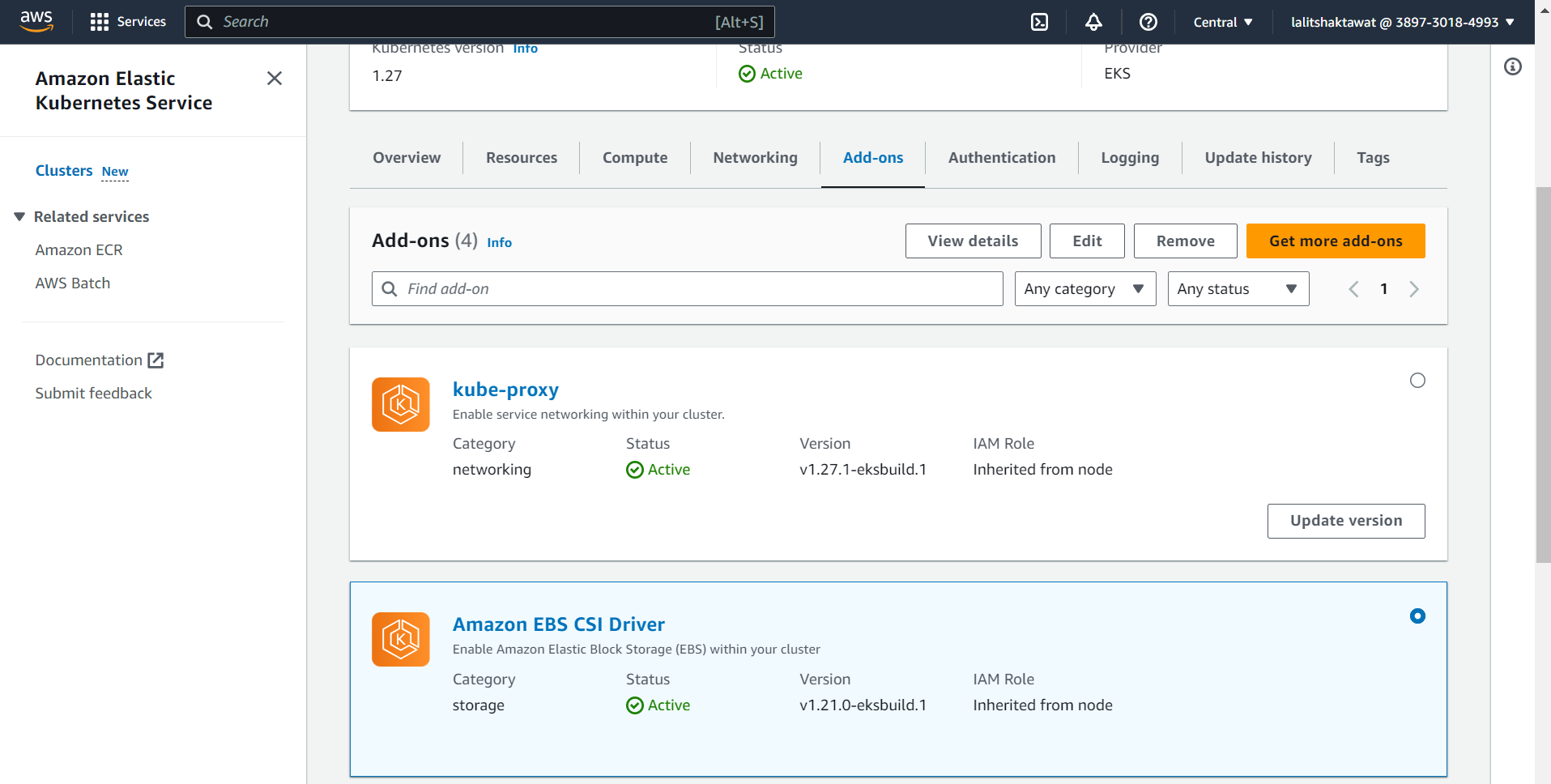
⭕ Update EBS CSI Driver fro IAM Role.
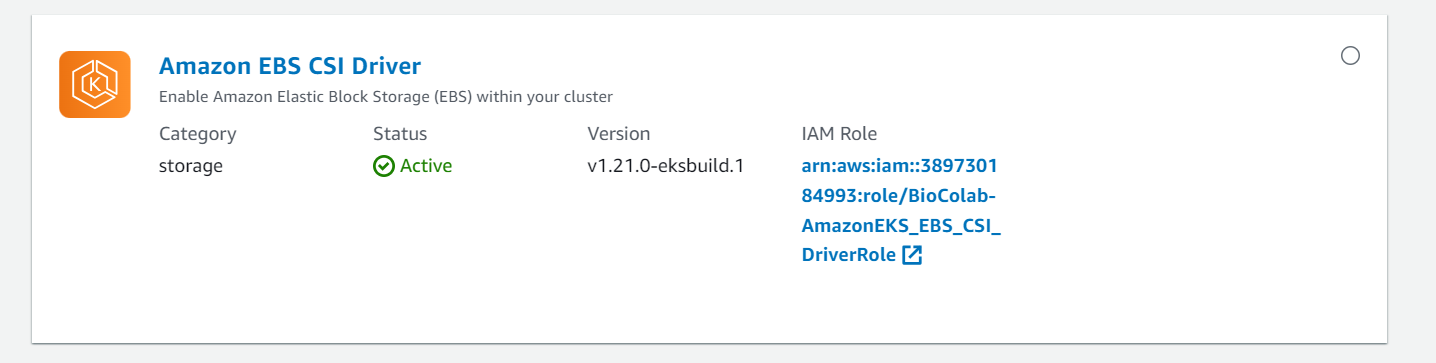
➡️ Test.
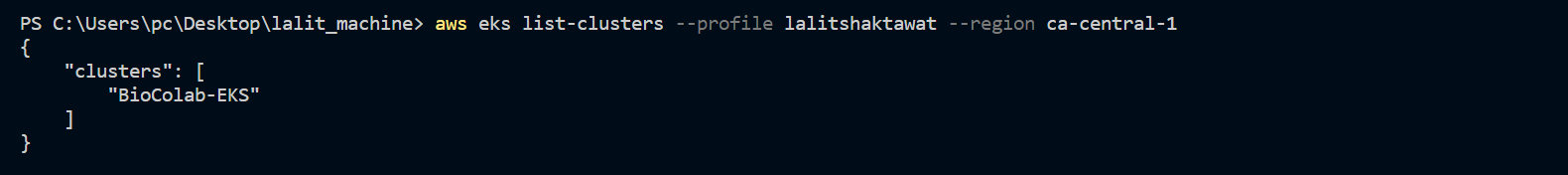
aws eks --region ca-central-1 describe-cluster --name BioColab-EKS --query cluster.status --profile lalitshaktawat
aws eks update-kubeconfig --region ca-central-1 --name BioColab-EKS --profile lalitshaktawat
eksctl utils associate-iam-oidc-provider --region=ca-central-1 --cluster=BioColab-EKS --approve --region ca-central-1 --profile lalitshaktawat
kubectl config current-context
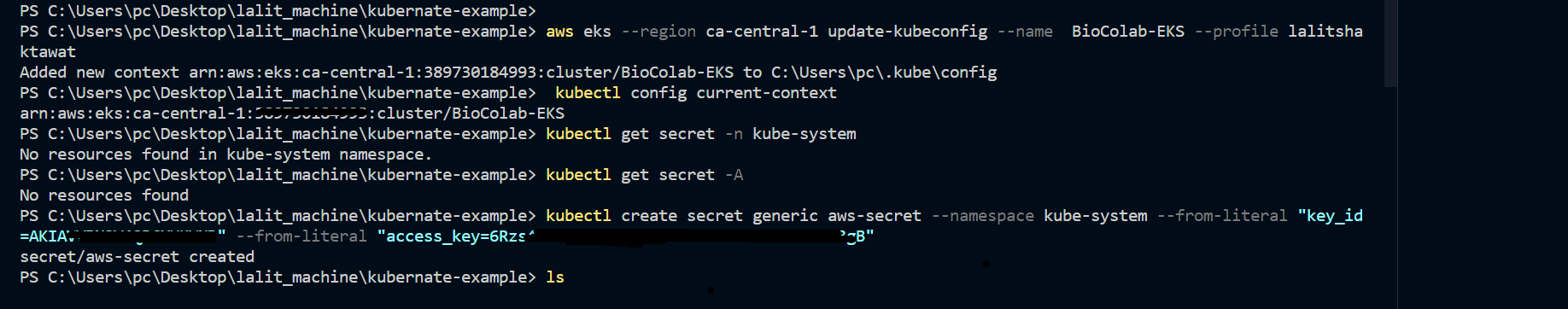
➡️ Troubleshoot.
We might face an issue related to PVC. Pod will not start and it will be on pending status. We just need to recreate ebs-csi-controller to resolve this issue.
kubectl delete pods -n kube-system -l=app=ebs-csi-controller

========
pvc.yaml
========
apiVersion: v1
kind: PersistentVolumeClaim
metadata:
name: ebs-claim1
spec:
accessModes:
- ReadWriteOnce
storageClassName: gp2
resources:
requests:
storage: 1Gi
========
pod.yaml
========
apiVersion: v1
kind: Pod
metadata:
name: app
spec:
containers:
- name: app
image: centos
command: ["/bin/sh"]
args: ["-c", "while true; do echo $(date -u) >> /data/out.txt; sleep 5; done"]
volumeMounts:
- name: persistent-storage
mountPath: /data
volumes:
- name: persistent-storage
persistentVolumeClaim:
claimName: ebs-claim1
Create resources for testing. It is just a test. You can skip this step.
========================================================================
kubectl apply -f .\pvc.yaml
kubectl apply -f .\pod.yaml
kubectl get pods
kubectl get pvc
Delete resources.
=================
kubectl delete po/<pod name> -n <namespace name>
⭕ Installing Helm.
➡️ Please follow link below to install Helm.
helm repo add bioturing https://bioturing.github.io/charts/apps/
helm repo update
helm search repo bioturing
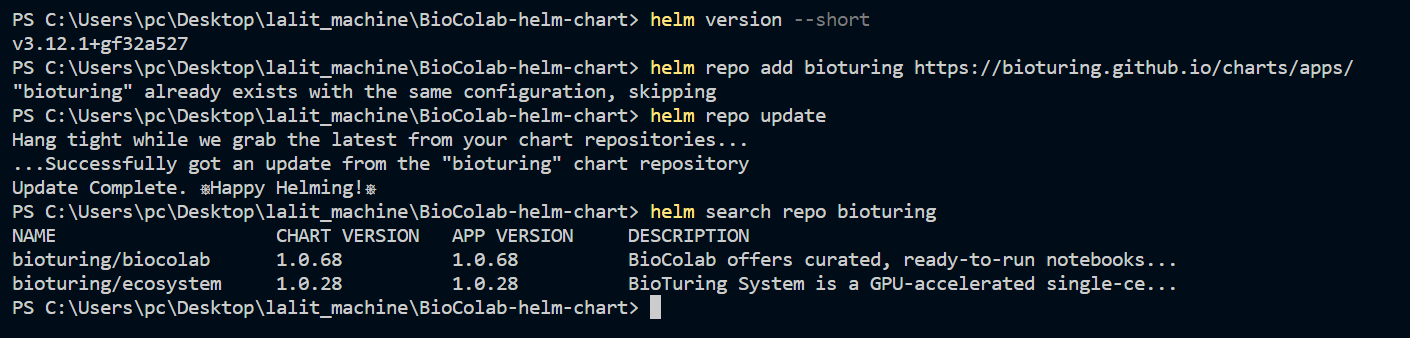
🔔 I downloaded latest version of BioStudio and adjusted values accordingly.
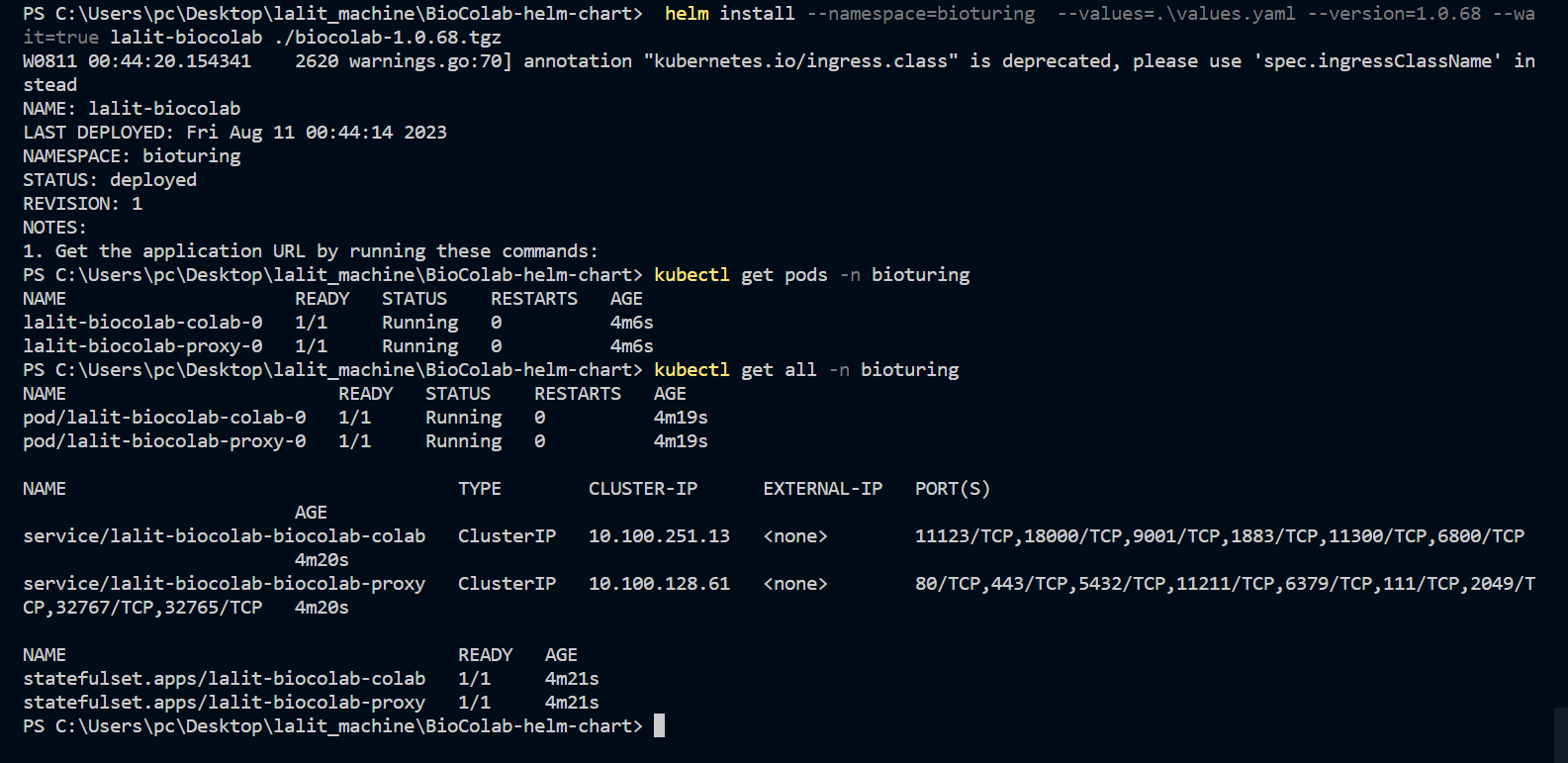
⭕ Kindly setup Load balancer (ALB) with your DNS.
⭕ Please contact 📧 [support@bioturing.com] for any inquiry.
Machine name on K8S
¶# We must use local DNS service name to add machine.
<Service Name>.<Namespace>.svc.cluster.local
# bioc-test-release-biocolab-colab.bioc-test.svc.cluster.local
# biocolab-preprod-biocolab-colab.bioturing-preprod.svc.cluster.local
BioStudio Checklist ¶
🔔 Note: Ensure that the BioProxy container doesn’t have HTTP_PROXY, HTTPS_PROXY, and NO_PROXY settings configured
1] Process running with host.
ps -ef
netstat -nltup
2] Process running with Bioproxy container
ps -ef
netstat -nltup
3] Process running with BioColab container
ps -ef
netstat -nltup
4] Check application up and running and connectivity.
curl command testing
curl <localhost>
curl <localhost>:<http port>
curl <localhost>:<application port>
curl <localhost>:<nginx port>
5] check whiltelist of domain with Host and both containers.
# curl https://colab.biotruing.com
Should show contents
# curl https://cdn.biotruing.com
# wget https://cdn.bioturing.com/documentation/adm.png
# curl https://cdn-eu-west-1.s3.eu-west-1.amazonaws.com
# wget https://cdn-eu-west-1.s3.eu-west-1.amazonaws.com/colab/apps/0w-byh0iNCWigGEjbZybU.92ae1dec-6bf9-4041-9d06-330e0fe7b564.zip
# curl https://s3.us-west-2.amazonaws.com/cdn.bioturing.com
# wget https://s3.us-west-2.amazonaws.com/cdn.bioturing.com/documentation/adm.png
# curl https://studio.bioturing.com
Should show contents
6] Check application varification processes
with Host machine
# ps -ef | grep 'docker'
# ps -ef | grep 'miniconda'
# ps -ef | grep nginx
# ps -ef | grep postgres
7] Check dashboard setting
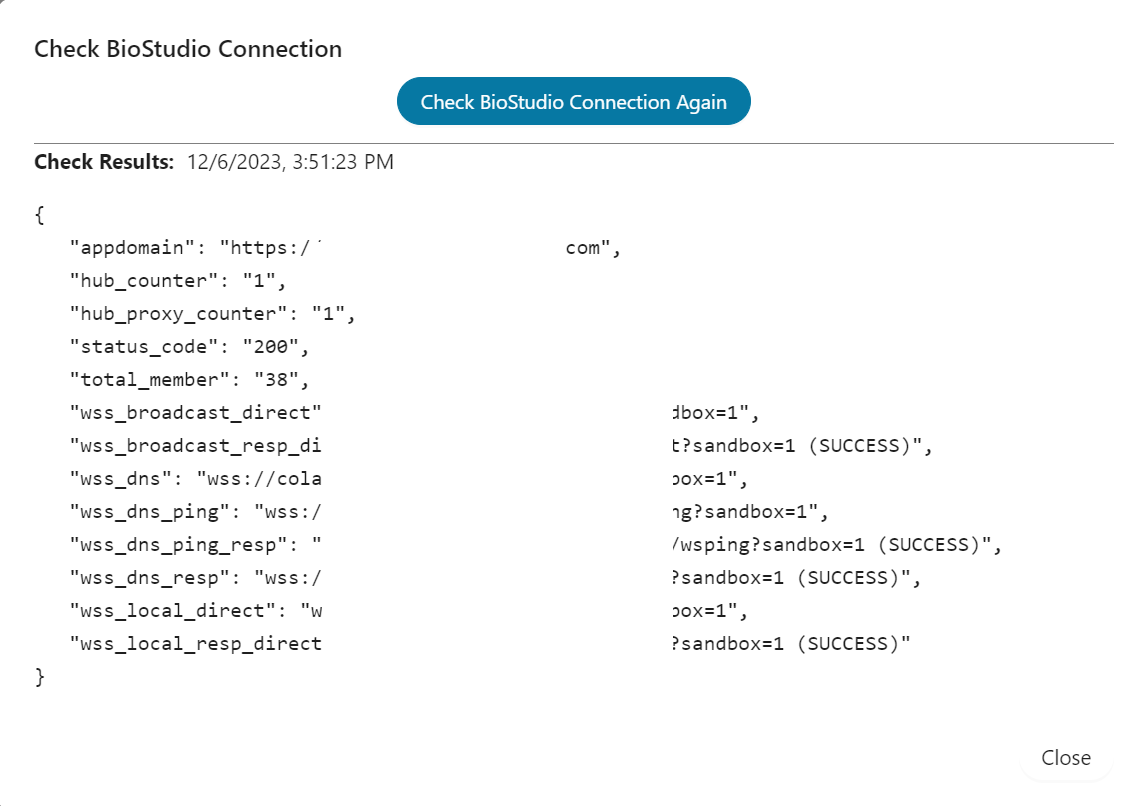
8] Check websocket
9] Check to download notebook and running
10] Guide for SSO login
Test and verification ¶
Before start installation below is the status:
root@ip-172-31-39-182:/biocolab/installation-2.1.11# netstat -nltup
Active Internet connections (only servers)
Proto Recv-Q Send-Q Local Address Foreign Address State PID/Program name
tcp 0 0 127.0.0.53:53 0.0.0.0:* LISTEN 2892/systemd-resolv
tcp 0 0 0.0.0.0:22 0.0.0.0:* LISTEN 2820/sshd: /usr/sbi
tcp6 0 0 :::22 :::* LISTEN 2820/sshd: /usr/sbi
udp 0 0 127.0.0.53:53 0.0.0.0:* 2892/systemd-resolv
udp 0 0 172.31.39.182:68 0.0.0.0:* 2882/systemd-networ
udp 0 0 127.0.0.1:323 0.0.0.0:* 2808/chronyd
udp6 0 0 ::1:323 :::* 2808/chronyd
root@ip-172-31-39-182:/biocolab/installation-2.1.11# env
SHELL=/bin/bash
PWD=/biocolab/installation-2.1.11
LOGNAME=root
HOME=/root
LANG=C.UTF-8
XDG_DATA_DIRS=/usr/local/share:/usr/share:/var/lib/snapd/desktop
PATH=/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin:/usr/games:/usr/local/games:/snap/bin
MAIL=/var/mail/root
_=/usr/bin/env
OLDPWD=/biocolab
Testing with normal structure ¶
After installation
⭕ HTTP port : 80 and HTTPS port : 443
Application port : 11123
netstat command testing ¶
Below are the port should be in as status.
1️⃣ Check netstat -nltup with Host and inside the container.
netstat -nltup # from host
root@ip-172-31-39-182:/biocolab/installation-2.1.11# netstat -nltup
Active Internet connections (only servers)
Proto Recv-Q Send-Q Local Address Foreign Address State PID/Program name
tcp 0 0 127.0.0.53:53 0.0.0.0:* LISTEN 2892/systemd-resolv
tcp 0 0 0.0.0.0:1883 0.0.0.0:* LISTEN 10842/docker-proxy
tcp 0 0 0.0.0.0:18000 0.0.0.0:* LISTEN 10736/docker-proxy
tcp 0 0 0.0.0.0:443 0.0.0.0:* LISTEN 10059/docker-proxy
tcp 0 0 0.0.0.0:80 0.0.0.0:* LISTEN 10081/docker-proxy
tcp 0 0 0.0.0.0:22 0.0.0.0:* LISTEN 2820/sshd: /usr/sbi
tcp 0 0 0.0.0.0:5432 0.0.0.0:* LISTEN 10009/docker-proxy
tcp 0 0 0.0.0.0:6800 0.0.0.0:* LISTEN 10821/docker-proxy
tcp 0 0 0.0.0.0:6379 0.0.0.0:* LISTEN 9982/docker-proxy
tcp 0 0 0.0.0.0:9091 0.0.0.0:* LISTEN 9958/docker-proxy
tcp 0 0 0.0.0.0:9001 0.0.0.0:* LISTEN 10802/docker-proxy
tcp 0 0 0.0.0.0:11300 0.0.0.0:* LISTEN 10757/docker-proxy
tcp 0 0 0.0.0.0:11211 0.0.0.0:* LISTEN 9937/docker-proxy
tcp 0 0 0.0.0.0:11123 0.0.0.0:* LISTEN 10778/docker-proxy
tcp 0 0 0.0.0.0:32767 0.0.0.0:* LISTEN 9893/docker-proxy
tcp 0 0 0.0.0.0:32765 0.0.0.0:* LISTEN 9915/docker-proxy
tcp6 0 0 :::1883 :::* LISTEN 10849/docker-proxy
tcp6 0 0 :::18000 :::* LISTEN 10744/docker-proxy
tcp6 0 0 :::443 :::* LISTEN 10066/docker-proxy
tcp6 0 0 :::80 :::* LISTEN 10088/docker-proxy
tcp6 0 0 :::22 :::* LISTEN 2820/sshd: /usr/sbi
tcp6 0 0 :::5432 :::* LISTEN 10030/docker-proxy
tcp6 0 0 :::6800 :::* LISTEN 10829/docker-proxy
tcp6 0 0 :::6379 :::* LISTEN 9988/docker-proxy
tcp6 0 0 :::9091 :::* LISTEN 9965/docker-proxy
tcp6 0 0 :::9001 :::* LISTEN 10809/docker-proxy
tcp6 0 0 :::11300 :::* LISTEN 10764/docker-proxy
tcp6 0 0 :::11211 :::* LISTEN 9943/docker-proxy
tcp6 0 0 :::11123 :::* LISTEN 10786/docker-proxy
tcp6 0 0 :::32767 :::* LISTEN 9900/docker-proxy
tcp6 0 0 :::32765 :::* LISTEN 9921/docker-proxy
udp 0 0 127.0.0.53:53 0.0.0.0:* 2892/systemd-resolv
udp 0 0 172.31.39.182:68 0.0.0.0:* 2882/systemd-networ
udp 0 0 127.0.0.1:323 0.0.0.0:* 2808/chronyd
udp6 0 0 ::1:323 :::* 2808/chronyd
netstat -nltup # inside the container
root@ip-172-31-39-182:/biocolab/installation-2.1.11# docker exec -it bioproxy /bin/bash
root@6c1fca69acf1:/home# netstat -nltup
Active Internet connections (only servers)
Proto Recv-Q Send-Q Local Address Foreign Address State PID/Program name
tcp 0 0 0.0.0.0:11211 0.0.0.0:* LISTEN -
tcp 0 0 127.0.0.1:8886 0.0.0.0:* LISTEN 183/node
tcp 0 0 127.0.0.1:9005 0.0.0.0:* LISTEN 167/python3
tcp 0 0 0.0.0.0:5432 0.0.0.0:* LISTEN -
tcp 0 0 0.0.0.0:6379 0.0.0.0:* LISTEN -
tcp 0 0 0.0.0.0:8077 0.0.0.0:* LISTEN 182/nginx: master p
tcp 0 0 127.0.0.1:5555 0.0.0.0:* LISTEN 298/dataplaneapi
tcp 0 0 0.0.0.0:80 0.0.0.0:* LISTEN -
tcp 0 0 0.0.0.0:443 0.0.0.0:* LISTEN -
tcp6 0 0 :::11211 :::* LISTEN -
tcp6 0 0 :::5432 :::* LISTEN -
tcp6 0 0 :::8077 :::* LISTEN 182/nginx: master p
udp 0 0 0.0.0.0:58951 0.0.0.0:* -
udp 0 0 0.0.0.0:36226 0.0.0.0:* -
root@6c1fca69acf1:/home#
-------
root@ip-172-31-39-182:/biocolab/installation-2.1.11# docker exec -it bioproxy /bin/bash
root@6c1fca69acf1:/home# netstat -nltup
Active Internet connections (only servers)
Proto Recv-Q Send-Q Local Address Foreign Address State PID/Program name
tcp 0 0 0.0.0.0:11211 0.0.0.0:* LISTEN -
tcp 0 0 127.0.0.1:8886 0.0.0.0:* LISTEN 183/node
tcp 0 0 127.0.0.1:9005 0.0.0.0:* LISTEN 167/python3
tcp 0 0 0.0.0.0:5432 0.0.0.0:* LISTEN -
tcp 0 0 0.0.0.0:6379 0.0.0.0:* LISTEN -
tcp 0 0 0.0.0.0:8077 0.0.0.0:* LISTEN 182/nginx: master p
tcp 0 0 127.0.0.1:5555 0.0.0.0:* LISTEN 298/dataplaneapi
tcp 0 0 0.0.0.0:80 0.0.0.0:* LISTEN -
tcp 0 0 0.0.0.0:443 0.0.0.0:* LISTEN -
tcp6 0 0 :::11211 :::* LISTEN -
tcp6 0 0 :::5432 :::* LISTEN -
tcp6 0 0 :::8077 :::* LISTEN 182/nginx: master p
udp 0 0 0.0.0.0:58951 0.0.0.0:* -
udp 0 0 0.0.0.0:36226 0.0.0.0:* -
root@6c1fca69acf1:/home#
-------
root@ip-172-31-39-182:/biocolab/installation-2.1.11# docker exec -it biocolab /bin/bash
root@5fc5db8bc5cf:/home# netstat -nltup
Active Internet connections (only servers)
Proto Recv-Q Send-Q Local Address Foreign Address State PID/Program name
tcp 0 0 0.0.0.0:9001 0.0.0.0:* LISTEN -
tcp 0 0 127.0.0.1:18001 0.0.0.0:* LISTEN 513/node
tcp 0 0 0.0.0.0:11123 0.0.0.0:* LISTEN 148/t2d_dsc_tool
tcp 0 0 0.0.0.0:11300 0.0.0.0:* LISTEN -
tcp 0 0 0.0.0.0:18081 0.0.0.0:* LISTEN 360/python3.10
tcp 0 0 0.0.0.0:18000 0.0.0.0:* LISTEN 513/node
tcp 0 0 0.0.0.0:1883 0.0.0.0:* LISTEN -
tcp 0 0 0.0.0.0:2223 0.0.0.0:* LISTEN 149/sshd: /usr/sbin
tcp 0 0 127.0.0.1:11113 0.0.0.0:* LISTEN 147/t2d_blc_tool
tcp 0 0 0.0.0.0:6800 0.0.0.0:* LISTEN 144/aria2c
tcp6 0 0 :::9001 :::* LISTEN -
tcp6 0 0 :::1883 :::* LISTEN -
tcp6 0 0 :::2222 :::* LISTEN 148/t2d_dsc_tool
tcp6 0 0 :::2223 :::* LISTEN 149/sshd: /usr/sbin
root@5fc5db8bc5cf:/home#
Environmental variables ¶
Bioproxy : environmental variable
# docker exec -it bioproxy /bin/bash
root@507cd2637a97:/home# env
NFS_T2_PORT=2049
SHELL=/bin/bash
POSTGRESQL_CLIENT_MIN_MESSAGES=error
DEBUG_MODE=false
HTTPS_SERVER_PORT=443
POSTGRESQL_USERNAME=XXXXXXXXXXXXXX
REDIS_PASSWORD=XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
root@507cd2637a97:/home#
root@507cd2637a97:/home# cat /etc/environment
root@507cd2637a97:/home#
root@507cd2637a97:/home# exit
BioColab : environmental variable
# docker exec -it biocolab /bin/bash
# env
root@c355340b52ce:/home# cat /etc/environment
PATH="/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin:/usr/games:/usr/local/games:/snap/bin"
no_proxy="kubernetes,.svc,.default,localhost,0.0.0.0,.cluster.local,100.10.0.0/16,fe80::/10,.local,100.10.0.3,fc00::/7,::1/128,182.168.10.0/24,10.0.0.0/8,10.82.0.90,127.0.0.1"
BioStudio process verification ¶
Process running with Bioproxy container
root@ip-172-31-39-123:/biocolab/installation-2.1.11/biocolab# docker exec -it bioproxy /bin/bash
root@507cd2637a97:/home# ps -ef
UID PID PPID C STIME TTY TIME CMD
root 1 0 0 03:50 pts/0 00:00:00 /bin/bash /super.sh
root 204 1 0 03:50 pts/0 00:00:02 /usr/bin/python3 /usr/bin/supervisord -n -c /etc/supervisor/supervisord.conf
root 205 204 0 03:50 pts/0 00:00:00 /usr/sbin/cron -f
memcach+ 206 204 0 03:50 pts/0 00:00:00 /usr/bin/memcached -p 11211 -u memcached -m 128 -c 1024000 -P /memcached/program.pid
postgres 207 204 0 03:50 pts/0 00:00:00 /opt/bitnami/postgresql/bin/postgres -D /bitnami/postgresql/data --config-file=/opt/bitnami/postgresql/conf/po
redis 208 204 0 03:50 pts/0 00:00:11 /usr/bin/redis-server 0.0.0.0:6379
root 219 204 0 03:50 pts/0 00:00:00 nginx: master process /usr/sbin/nginx -g daemon off;
root 220 204 0 03:50 pts/0 00:00:00 /usr/bin/node /server.js
www-data 221 204 0 03:50 pts/0 00:00:04 /usr/local/sbin/haproxy -f /etc/haproxy/haproxy.cfg -p /var/run/haproxy.pid
www-data 253 219 0 03:50 pts/0 00:00:00 nginx: worker process
www-data 254 219 0 03:50 pts/0 00:00:00 nginx: worker process
www-data 255 219 0 03:50 pts/0 00:00:00 nginx: worker process
www-data 257 219 0 03:50 pts/0 00:00:00 nginx: worker process
postgres 308 207 0 03:50 ? 00:00:00 postgres: checkpointer
postgres 309 207 0 03:50 ? 00:00:00 postgres: background writer
postgres 311 207 0 03:50 ? 00:00:00 postgres: walwriter
postgres 312 207 0 03:50 ? 00:00:00 postgres: autovacuum launcher
postgres 313 207 0 03:50 ? 00:00:00 postgres: logical replication launcher
root 336 204 0 03:50 pts/0 00:00:02 /usr/local/bin/dataplaneapi --port 5555 -b /usr/local/sbin/haproxy -c /etc/haproxy/haproxy.cfg -d 5 -r /usr/bi
postgres 705 207 0 03:58 ? 00:00:00 postgres: postgres postgres 172.17.0.1(37132) idle
postgres 706 207 0 03:58 ? 00:00:00 postgres: postgres biocolab 172.17.0.1(37140) idle
postgres 707 207 0 03:58 ? 00:00:00 postgres: postgres biocolab 172.17.0.1(37156) idle
postgres 1036 207 0 04:08 ? 00:00:00 postgres: postgres biocohub 172.17.0.1(57498) idle
root 4206 0 1 05:33 pts/1 00:00:00 /bin/bash
root 4212 4206 0 05:33 pts/1 00:00:00 ps -ef
root@507cd2637a97:/home#
Process running with BioColab container
root@ip-172-31-39-123:/biocolab/installation-2.1.11/biocolab# docker exec -it biocolab /bin/bash
root@b1a226ccc20a:/home# ps -ef
UID PID PPID C STIME TTY TIME CMD
root 1 0 0 03:56 pts/0 00:00:00 /bin/bash /super.sh
root 110 1 0 03:56 pts/0 00:00:09 /usr/bin/python3 /usr/bin/supervisord -n -c /etc/supervisor/supervisord.conf
mosquit+ 112 110 0 03:56 pts/0 00:00:02 /usr/local/sbin/mosquitto -c /mosquitto/config/mosquitto.conf
beansta+ 113 110 0 03:56 pts/0 00:00:00 /usr/local/bin/beanstalkd -b /appdata/beanstalkd -f 1000 -u beanstalkd
root 115 110 0 03:56 pts/0 00:00:00 /usr/bin/Xvfb :0.0 -screen 0 800x600x16 -dpi 75 -nolisten tcp -audit 4 -ac -auth /root/.Xauthority
root 125 110 0 03:56 pts/0 00:00:00 /usr/bin/aria2c --enable-rpc --rpc-listen-all --rpc-allow-origin-all --rpc-secret=************ --rpc-listen-po
root 128 110 0 03:56 pts/0 00:00:00 sshd: /usr/sbin/sshd -p 2223 -D [listener] 0 of 10-100 startups
root 129 110 0 03:56 pts/0 00:00:00 /usr/sbin/cron -f
root 462 110 1 03:58 pts/0 00:01:43 /appdata/apps/t2d_dsc_tool
root 484 110 0 03:58 pts/0 00:00:10 /appdata/apps/t2d_blc_tool
root 1846 110 0 04:06 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 1858 110 0 04:06 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 1867 110 0 04:06 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 1880 110 0 04:06 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 1881 110 0 04:06 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 1882 110 0 04:06 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 1883 110 0 04:06 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 1884 110 0 04:06 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 1885 110 0 04:06 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 1886 110 0 04:06 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 1887 110 0 04:06 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 1888 110 0 04:06 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 1889 110 0 04:06 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 1890 110 0 04:06 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 1891 110 0 04:06 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 1957 110 0 04:08 pts/0 00:00:00 /bin/bash /start.sh
root 1958 1957 0 04:08 pts/0 00:00:04 /miniconda/user/bin/python3.10 /miniconda/user/bin/jupyterhub --config /configs/hub/etc/config.py --ip 0.0.0.0
root 1965 1958 0 04:08 ? 00:00:01 node /miniconda/user/bin/configurable-http-proxy --ip 0.0.0.0 --port 18000 --api-ip 127.0.0.1 --api-port 18001
root 1975 1958 0 04:08 ? 00:00:00 /miniconda/user/bin/python3.10 -m jupyterhub_idle_culler --timeout=36000
root 2021 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2022 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2023 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2024 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2025 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2026 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2027 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2028 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2029 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2030 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2031 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2032 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2033 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2034 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2035 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2036 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2037 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2038 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2039 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2040 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 2094 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-pack-notebook-worker
root 2095 110 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-pack-notebook-worker
root 2107 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2108 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2109 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2110 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2111 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2112 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2113 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2114 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2115 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2116 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2117 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2118 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2119 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2120 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2121 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2122 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2123 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2124 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2125 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2126 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2127 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2128 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2129 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2130 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 2131 110 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 5607 0 1 05:34 pts/1 00:00:00 /bin/bash
root 5619 5607 0 05:34 pts/1 00:00:00 ps -ef
root@b1a226ccc20a:/home#
curl command testing ¶
curl <localhost>
curl <localhost>:<http port>
curl <localhost>:<application port>
curl <localhost>:<nginx port>
🔔 NOTE : BioStudio is running on http (80) and https (443)
Curl with host
root@ip-172-31-39-182:/biocolab/installation-2.1.11# curl localhost
<html lang="en">
<head>
<meta charset="UTF-8">
<meta http-equiv="X-UA-Compatible" content="IE=edge">
<meta name="viewport" content="width=device-width, initial-scale=1.0">
<meta property="og:type" content="article" />
<meta property="og:url" content="https://studio.bioturing.com/" />
<meta property="og:description" content="BioStudio is the complete
-------
-------
root@ip-172-31-39-182:/biocolab/installation-2.1.11# curl localhost:80
<head>
<meta charset="UTF-8">
<meta http-equiv="X-UA-Compatible" content="IE=edge">
<meta name="viewport" content="width=device-width, initial-scale=1.0">
<meta property="og:type" content="article" />
<meta property="og:url" content="https://studio.bioturing.com/" />
<meta property="og:description" content="BioStudio is the complete
-------
-------
root@ip-172-31-39-182:/biocolab/installation-2.1.11# curl localhost:11123
<head>
<meta charset="UTF-8">
<meta http-equiv="X-UA-Compatible" content="IE=edge">
<meta name="viewport" content="width=device-width, initial-scale=1.0">
<meta property="og:type" content="article" />
<meta property="og:url" content="https://studio.bioturing.com/" />
<meta property="og:description" content="BioStudio is the complete
Curl inside the container
# BioProxy container
root@ip-172-31-39-182:/biocolab/installation-2.1.11# docker exec -it bioproxy /bin/bash
root@6c1fca69acf1:/home# curl localhost
<html lang="en">
<head>
<meta charset="UTF-8">
<meta http-equiv="X-UA-Compatible" content="IE=edge">
<meta name="viewport" content="width=device-width, initial-scale=1.0">
<meta property="og:type" content="article" />
<meta property="og:url" content="https://studio.bioturing.com/" />
<meta property="og:description" content="BioStudio is the complete analytics platform that empowers scientists to expand their research
horizons without technical complexities." />
-------
-------
# BioColab Container
NOTE: Here we can only curl with application port.
root@5fc5db8bc5cf:/home# curl localhost:11123
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
37 168k 37 65219 <!DOCTYPE html> 0 --:--:-- --:--:-- --:--:-- 0
0 <html lang="en">
<head>
0 159k <meta charset="UTF-8">
<meta http-equiv="X-UA-Compatible" content="IE=edge">
<meta name="viewport" content="width=device-width, initial-scale=1.0">
Check nslookup ¶
nslookup <Domain name>
root@ip-172-31-39-182:/biocolab/installation-2.1.11# nslookup <Client BioStudio Domain>
Server: 127.0.0.53
Address: 127.0.0.53#53
Non-authoritative answer:
Name: <Client BioStudio Domain>
Address: 54.203.5.109
root@ip-172-31-39-182:/biocolab/installation-2.1.11#
check whiltelist of domian ¶
Check with host
# curl https://colab.biotruing.com
-- Should show contents
# curl https://cdn.biotruing.com
# wget https://cdn.bioturing.com/documentation/adm.png
# curl https://cdn-eu-west-1.s3.eu-west-1.amazonaws.com
# wget https://cdn-eu-west-1.s3.eu-west-1.amazonaws.com/colab/apps/0w-byh0iNCWigGEjbZybU.92ae1dec-6bf9-4041-9d06-330e0fe7b564.zip
# curl https://s3.us-west-2.amazonaws.com/cdn.bioturing.com
# wget https://s3.us-west-2.amazonaws.com/cdn.bioturing.com/documentation/adm.png
# curl https://studio.bioturing.com
-- Should show contents
Check inside the both containers
# Both container should show the same output and all would have the result.
# curl https://colab.biotruing.com
-- Should show contents
# curl https://cdn.biotruing.com
# wget https://cdn.bioturing.com/documentation/adm.png
# curl https://cdn-eu-west-1.s3.eu-west-1.amazonaws.com
# wget https://cdn-eu-west-1.s3.eu-west-1.amazonaws.com/colab/apps/0w-byh0iNCWigGEjbZybU.92ae1dec-6bf9-4041-9d06-330e0fe7b564.zip
# curl https://s3.us-west-2.amazonaws.com/cdn.bioturing.com
# wget https://s3.us-west-2.amazonaws.com/cdn.bioturing.com/documentation/adm.png
# curl https://studio.bioturing.com
-- Should show contents
Check application varification processes ¶
with Host machine
# ps -ef | grep 'docker'
# ps -ef | grep 'miniconda'
# ps -ef | grep nginx
# ps -ef | grep postgres
root@ip-172-31-39-182:/biocolab/installation-2.1.11/test# ps -ef | grep 'docker'
root 4243 1 1 03:24 ? 00:02:54 /usr/bin/dockerd -H fd:// --containerd=/run/containerd/containerd.sock
root 9893 4243 0 04:04 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip 0.0.0.0 -host-port 32767 -container-ip 172.17.0.2 -container-port 32767
root 9900 4243 0 04:04 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip :: -host-port 32767 -container-ip 172.17.0.2 -container-port 32767
root 9915 4243 0 04:04 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip 0.0.0.0 -host-port 32765 -container-ip 172.17.0.2 -container-port 32765
root 9921 4243 0 04:04 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip :: -host-port 32765 -container-ip 172.17.0.2 -container-port 32765
root 9937 4243 0 04:04 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip 0.0.0.0 -host-port 11211 -container-ip 172.17.0.2 -container-port 11211
root 9943 4243 0 04:04 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip :: -host-port 11211 -container-ip 172.17.0.2 -container-port 11211
root 9958 4243 0 04:04 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip 0.0.0.0 -host-port 9091 -container-ip 172.17.0.2 -container-port 9091
root 9965 4243 0 04:04 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip :: -host-port 9091 -container-ip 172.17.0.2 -container-port 9091
root 9982 4243 0 04:04 ? 00:00:06 /usr/bin/docker-proxy -proto tcp -host-ip 0.0.0.0 -host-port 6379 -container-ip 172.17.0.2 -container-port 6379
root 9988 4243 0 04:04 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip :: -host-port 6379 -container-ip 172.17.0.2 -container-port 6379
root 10009 4243 0 04:04 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip 0.0.0.0 -host-port 5432 -container-ip 172.17.0.2 -container-port 5432
root 10030 4243 0 04:04 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip :: -host-port 5432 -container-ip 172.17.0.2 -container-port 5432
root 10059 4243 0 04:04 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip 0.0.0.0 -host-port 443 -container-ip 172.17.0.2 -container-port 443
root 10066 4243 0 04:04 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip :: -host-port 443 -container-ip 172.17.0.2 -container-port 443
root 10081 4243 0 04:04 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip 0.0.0.0 -host-port 80 -container-ip 172.17.0.2 -container-port 80
root 10088 4243 0 04:04 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip :: -host-port 80 -container-ip 172.17.0.2 -container-port 80
root 10736 4243 0 04:08 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip 0.0.0.0 -host-port 18000 -container-ip 172.17.0.3 -container-port 18000
root 10744 4243 0 04:08 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip :: -host-port 18000 -container-ip 172.17.0.3 -container-port 18000
root 10757 4243 0 04:08 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip 0.0.0.0 -host-port 11300 -container-ip 172.17.0.3 -container-port 11300
root 10764 4243 0 04:08 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip :: -host-port 11300 -container-ip 172.17.0.3 -container-port 11300
root 10778 4243 0 04:08 ? 00:00:05 /usr/bin/docker-proxy -proto tcp -host-ip 0.0.0.0 -host-port 11123 -container-ip 172.17.0.3 -container-port 11123
root 10786 4243 0 04:08 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip :: -host-port 11123 -container-ip 172.17.0.3 -container-port 11123
root 10802 4243 0 04:08 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip 0.0.0.0 -host-port 9001 -container-ip 172.17.0.3 -container-port 9001
root 10809 4243 0 04:08 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip :: -host-port 9001 -container-ip 172.17.0.3 -container-port 9001
root 10821 4243 0 04:08 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip 0.0.0.0 -host-port 6800 -container-ip 172.17.0.3 -container-port 6800
root 10829 4243 0 04:08 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip :: -host-port 6800 -container-ip 172.17.0.3 -container-port 6800
root 10842 4243 0 04:08 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip 0.0.0.0 -host-port 1883 -container-ip 172.17.0.3 -container-port 1883
root 10849 4243 0 04:08 ? 00:00:00 /usr/bin/docker-proxy -proto tcp -host-ip :: -host-port 1883 -container-ip 172.17.0.3 -container-port 1883
root 46489 1507 0 07:54 pts/1 00:00:00 grep --color=auto docker
root@ip-172-31-39-182:/biocolab/installation-2.1.11/test#
-------
-------
root@ip-172-31-39-182:/biocolab/installation-2.1.11/test# ps -ef | grep 'miniconda'
root 11186 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11187 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11188 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11189 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11190 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11191 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11192 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11193 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11194 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11195 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11196 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11197 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11199 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11200 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11201 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11202 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11203 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11204 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11205 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11206 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 11218 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-pack-notebook-worker
root 11219 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-pack-notebook-worker
root 11220 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11221 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11222 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11223 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11224 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11225 11035 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11226 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11229 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11230 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11231 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11232 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11234 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11235 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11236 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11237 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11238 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11239 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11240 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11241 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11242 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11243 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11244 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11245 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11246 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11247 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 11248 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 11249 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 11250 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 11251 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 11252 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 11253 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 11254 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 11255 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 11256 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 11257 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 11258 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 11259 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 11260 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 11261 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 11262 11035 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 11264 11263 0 04:08 pts/0 00:00:10 /miniconda/user/bin/python3.10 /miniconda/user/bin/jupyterhub --config /configs/hub/etc/config.py --ip 0.0.0.0 --port 18000 --no-ssl
root 11422 11264 0 04:08 ? 00:00:02 node /miniconda/user/bin/configurable-http-proxy --ip 0.0.0.0 --port 18000 --api-ip 127.0.0.1 --api-port 18001 --error-target http://5fc5db8bc5cf:18081/hub/error --log-level info
root 11436 11264 0 04:08 ? 00:00:00 /miniconda/user/bin/python3.10 -m jupyterhub_idle_culler --timeout=36000
root 46573 1507 0 07:54 pts/1 00:00:00 grep --color=auto miniconda
root@ip-172-31-39-182:/biocolab/installation-2.1.11/test#
-------
-------
root@ip-172-31-39-182:/biocolab/installation-2.1.11/test# ps -ef | grep nginx
root 10327 10312 0 04:04 pts/0 00:00:00 nginx: master process /usr/sbin/nginx -g daemon off;
www-data 10374 10327 0 04:04 pts/0 00:00:00 nginx: worker process
www-data 10375 10327 0 04:04 pts/0 00:00:00 nginx: worker process
www-data 10376 10327 0 04:04 pts/0 00:00:00 nginx: worker process
www-data 10377 10327 0 04:04 pts/0 00:00:00 nginx: worker process
root 46627 1507 0 07:54 pts/1 00:00:00 grep --color=auto nginx
root@ip-172-31-39-182:/biocolab/installation-2.1.11/test#
-------
-------
root@ip-172-31-39-182:/biocolab/installation-2.1.11/test# ps -ef | grep postgres
ubuntu 10315 10312 0 04:04 pts/0 00:00:00 /opt/bitnami/postgresql/bin/postgres -D /bitnami/postgresql/data --config-file=/opt/bitnami/postgresql/conf/postgresql.conf --external_pid_file=/opt/bitnami/postgresql/tmp/postgresql.pid --hba_file=/opt/bitnami/postgresql/conf/pg_hba.conf
ubuntu 10384 10315 0 04:04 ? 00:00:00 postgres: checkpointer
ubuntu 10385 10315 0 04:04 ? 00:00:00 postgres: background writer
ubuntu 10388 10315 0 04:04 ? 00:00:00 postgres: walwriter
ubuntu 10389 10315 0 04:04 ? 00:00:00 postgres: autovacuum launcher
ubuntu 10390 10315 0 04:04 ? 00:00:00 postgres: logical replication launcher
ubuntu 11335 10315 0 04:08 ? 00:00:00 postgres: postgres postgres 172.17.0.1(38918) idle
ubuntu 11336 10315 0 04:08 ? 00:00:00 postgres: postgres biocolab 172.17.0.1(38924) idle
ubuntu 11421 10315 0 04:08 ? 00:00:00 postgres: postgres biocohub 172.17.0.1(38936) idle
ubuntu 11516 10315 0 04:09 ? 00:00:00 postgres: postgres biocolab 172.17.0.1(55506) idle
root 46638 1507 0 07:55 pts/1 00:00:00 grep --color=auto postgres
root@ip-172-31-39-182:/biocolab/installation-2.1.11/test#
Check process with BioProxy container
root@ip-172-31-39-182:/biocolab/installation-2.1.11/test# docker exec -it bioproxy /bin/bash
root@6c1fca69acf1:/home# ps -ef
UID PID PPID C STIME TTY TIME CMD
root 1 0 0 04:04 pts/0 00:00:00 /bin/bash /super.sh
root 167 1 0 04:04 pts/0 00:00:06 /usr/bin/python3 /usr/bin/supervisord -n -c /etc/supervisor/supervisord.conf
root 168 167 0 04:04 pts/0 00:00:00 /usr/sbin/cron -f
memcach+ 169 167 0 04:04 pts/0 00:00:02 /usr/bin/memcached -p 11211 -u memcached -m 128 -c 1024000 -P /memcached/program.pid
postgres 170 167 0 04:04 pts/0 00:00:00 /opt/bitnami/postgresql/bin/postgres -D /bitnami/postgresql/data --config-file=/opt/bit
redis 171 167 0 04:04 pts/0 00:00:25 /usr/bin/redis-server 0.0.0.0:6379
root 182 167 0 04:04 pts/0 00:00:00 nginx: master process /usr/sbin/nginx -g daemon off;
root 183 167 0 04:04 pts/0 00:00:00 /usr/bin/node /server.js
www-data 184 167 0 04:04 pts/0 00:00:10 /usr/local/sbin/haproxy -f /etc/haproxy/haproxy.cfg -p /var/run/haproxy.pid
www-data 229 182 0 04:04 pts/0 00:00:00 nginx: worker process
www-data 230 182 0 04:04 pts/0 00:00:00 nginx: worker process
www-data 231 182 0 04:04 pts/0 00:00:00 nginx: worker process
www-data 232 182 0 04:04 pts/0 00:00:00 nginx: worker process
postgres 239 170 0 04:04 ? 00:00:00 postgres: checkpointer
postgres 240 170 0 04:04 ? 00:00:00 postgres: background writer
postgres 243 170 0 04:04 ? 00:00:00 postgres: walwriter
postgres 244 170 0 04:04 ? 00:00:00 postgres: autovacuum launcher
postgres 245 170 0 04:04 ? 00:00:00 postgres: logical replication launcher
root 298 167 0 04:04 pts/0 00:00:05 /usr/local/bin/dataplaneapi --port 5555 -b /usr/local/sbin/haproxy -c /etc/haproxy/hapr
postgres 467 170 0 04:08 ? 00:00:00 postgres: postgres postgres 172.17.0.1(38918) idle
postgres 468 170 0 04:08 ? 00:00:00 postgres: postgres biocolab 172.17.0.1(38924) idle
postgres 469 170 0 04:08 ? 00:00:00 postgres: postgres biocohub 172.17.0.1(38936) idle
postgres 473 170 0 04:09 ? 00:00:00 postgres: postgres biocolab 172.17.0.1(55506) idle
root 9008 0 0 07:57 pts/1 00:00:00 /bin/bash
root 9014 9008 0 07:57 pts/1 00:00:00 ps -ef
root@6c1fca69acf1:/home#
Check process with BioColab container
root@5fc5db8bc5cf:/home# ps -ef
UID PID PPID C STIME TTY TIME CMD
root 1 0 0 04:08 pts/0 00:00:00 /bin/bash /super.sh
root 131 1 0 04:08 pts/0 00:00:10 /usr/bin/python3 /usr/bin/supervisord -n -c /etc/supervisor/supervisord.conf
mosquit+ 133 131 0 04:08 pts/0 00:00:06 /usr/local/sbin/mosquitto -c /mosquitto/config/mosquitto.conf
beansta+ 134 131 0 04:08 pts/0 00:00:00 /usr/local/bin/beanstalkd -b /appdata/beanstalkd -f 1000 -u beanstalkd
root 136 131 0 04:08 pts/0 00:00:00 /usr/bin/Xvfb :0.0 -screen 0 800x600x16 -dpi 75 -nolisten tcp -audit 4 -ac -auth /root/
root 144 131 0 04:08 pts/0 00:00:00 /usr/bin/aria2c --enable-rpc --rpc-listen-all --rpc-allow-origin-all --rpc-secret=*****
root 147 131 0 04:08 pts/0 00:00:22 /appdata/apps/t2d_blc_tool
root 148 131 0 04:08 pts/0 00:01:50 /appdata/apps/t2d_dsc_tool
root 149 131 0 04:08 pts/0 00:00:00 sshd: /usr/sbin/sshd -p 2223 -D [listener] 0 of 10-100 startups
root 150 131 0 04:08 pts/0 00:00:00 /usr/sbin/cron -f
root 282 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 283 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 284 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 285 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 286 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 287 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 288 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 289 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 290 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 291 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 292 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 293 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 295 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 296 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 297 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 298 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 299 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 300 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 301 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 302 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-custom-task-worker
root 314 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-pack-notebook-worker
root 315 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-pack-notebook-worker
root 316 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 317 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 318 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 319 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 320 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 321 131 0 04:08 pts/0 00:00:00 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 322 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 325 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 326 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 327 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 328 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 330 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 331 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 332 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 333 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 334 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 335 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 336 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 337 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 338 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 339 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 340 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 341 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 342 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 343 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-setup-notebook-worker
root 344 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 345 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 346 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 347 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 348 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 349 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 350 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 351 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 352 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 353 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 354 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 355 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 356 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 357 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 358 131 0 04:08 pts/0 00:00:01 /miniconda/user/bin/python3.10 /miniconda/user/bin/bhub-create-kernel-worker
root 359 131 0 04:08 pts/0 00:00:00 /bin/bash /start.sh
root 360 359 0 04:08 pts/0 00:00:10 /miniconda/user/bin/python3.10 /miniconda/user/bin/jupyterhub --config /configs/hub/etc
root 513 360 0 04:08 ? 00:00:02 node /miniconda/user/bin/configurable-http-proxy --ip 0.0.0.0 --port 18000 --api-ip 127
root 527 360 0 04:08 ? 00:00:00 /miniconda/user/bin/python3.10 -m jupyterhub_idle_culler --timeout=36000
root 9394 0 0 07:58 pts/1 00:00:00 /bin/bash
root 9406 9394 0 07:58 pts/1 00:00:00 ps -ef
root@5fc5db8bc5cf:/home#
Application location ¶
Application location
BioColab
docker exec -it biocolab /bin/bash
root@5fc5db8bc5cf:/appdata/share/miniconda# ll
total 16
drwxr-xr-x 4 root root 4096 Dec 15 04:08 ./
drwxr-xr-x 6 root root 4096 Dec 15 03:36 ../
drwxr-xr-x 2 root root 4096 Sep 18 08:09 apps/
lrwxrwxrwx 1 root root 24 Dec 15 04:08 miniconda -> /appdata/share/miniconda/
drwxr-xr-x 38 root root 4096 Oct 10 12:41 user/
root@5fc5db8bc5cf:/appdata/share/miniconda# pwd
/appdata/share/miniconda
root@5fc5db8bc5cf:/appdata/share/miniconda#
Installation for APP
root@5fc5db8bc5cf:/appdata/.bbcache# ls -l
total 12
-rwx------ 1 root root 908 Dec 15 08:08 kjjsdkfjslkdmlkclriltrij78783749879875.license
-rw------- 1 root root 908 Dec 15 08:08 kjjsdkfjslkdmlkclriltrij78783749879875.license_bk
drwx------ 4 root root 4096 Dec 15 03:30 searching
root@5fc5db8bc5cf:/appdata/.bbcache# ls searching/
conda.bleve download
root@5fc5db8bc5cf:/appdata/.bbcache# tree
.
├── kjjsdkfjslkdmlkclriltrij78783749879875.license
├── kjjsdkfjslkdmlkclriltrij78783749879875.license_bk
└── searching
├── conda.bleve
│ ├── index_meta.json
│ └── store
│ ├── 00000000013c.zap
│ ├── 00000000014f.zap
│ ├── 000000000155.zap
│ ├── 000000000164.zap
│ ├── 000000000173.zap
│ ├── 00000000017d.zap
│ ├── 000000000191.zap
│ ├── 000000000192.zap
│ └── root.bolt
└── download
├── anaconda_channeldata.json
├── anaconda_channeldata.json.converted
├── anaconda_linux-64.json
├── anaconda_noarch.json
├── bioconda_linux-64.json
├── bioconda_noarch.json
├── bioturing_linux-64.json
├── bioturing_noarch.json
├── cctbx202211_linux-64.json
├── cctbx202211_noarch.json
├── conda-forge_channeldata.json
├── conda-forge_channeldata.json.converted
├── conda-forge_linux-64.json
├── conda-forge_noarch.json
├── fastai_linux-64.json
├── fastai_noarch.json
├── fastchan_linux-64.json
├── fastchan_noarch.json
├── intel_linux-64.json
├── intel_noarch.json
├── main_linux-64.json
├── main_noarch.json
├── nvidia_linux-64.json
├── nvidia_noarch.json
├── ome_linux-64.json
├── ome_noarch.json
├── prometeia_linux-64.json
├── prometeia_noarch.json
├── pytorch_linux-64.json
├── pytorch_noarch.json
├── r_linux-64.json
├── r_noarch.json
├── sunpy_linux-64.json
└── sunpy_noarch.json
4 directories, 46 files
root@5fc5db8bc5cf:/appdata/.bbcache#
Application log
root@5fc5db8bc5cf:/appdata/logs# pwd
/appdata/logs
root@5fc5db8bc5cf:/appdata/logs# ls -l
total 0
-rwx------ 1 root root 0 Dec 15 03:44 jupyterhub.log
root@5fc5db8bc5cf:/appdata/logs#
# ls -la /var/log/supervisor
root@5fc5db8bc5cf:/appdata/logs# ls -la /var/log/supervisor
total 444
drwx------ 1 root root 4096 Dec 15 04:08 .
drwxr-xr-x 1 root root 4096 Dec 15 04:08 ..
-rwx------ 1 root root 0 Dec 15 04:08 aria2c_stderr.log
-rwx------ 1 root root 12536 Dec 15 07:33 aria2c_stdout.log
-rwx------ 1 root root 0 Dec 15 04:08 beanstalkd_stderr.log
-rwx------ 1 root root 0 Dec 15 04:08 beanstalkd_stdout.log
-rw-r--r-- 1 root root 0 Dec 15 04:08 bhub-create-custom-task-worker_stderr.log
-rw-r--r-- 1 root root 108160 Dec 15 08:03 bhub-create-custom-task-worker_stdout.log
-rw-r--r-- 1 root root 0 Dec 15 04:08 bhub-create-kernel-worker_stderr.log
-rw-r--r-- 1 root root 90120 Dec 15 08:03 bhub-create-kernel-worker_stdout.log
-rw-r--r-- 1 root root 0 Dec 15 04:08 bhub-pack-notebook-worker_stderr.log
-rw-r--r-- 1 root root 11416 Dec 15 08:03 bhub-pack-notebook-worker_stdout.log
-rw-r--r-- 1 root root 0 Dec 15 04:08 bhub-setup-notebook-worker_stderr.log
-rw-r--r-- 1 root root 143950 Dec 15 08:03 bhub-setup-notebook-worker_stdout.log
-rwx------ 1 root root 0 Dec 15 04:08 cleanup_stderr.log
-rwx------ 1 root root 24 Dec 15 04:08 cleanup_stdout.log
-rwx------ 1 root root 0 Dec 15 04:08 colab-unset-vars_stderr.log
-rwx------ 1 root root 0 Dec 15 04:08 colab-unset-vars_stdout.log
-rwx------ 1 root root 720 Dec 15 04:08 colab_stderr.log
-rwx------ 1 root root 151 Dec 15 04:08 colab_stdout.log
-rwx------ 1 root root 4050 Dec 15 04:08 colabblc_stderr.log
-rwx------ 1 root root 0 Dec 15 04:08 colabblc_stdout.log
-rwx------ 1 root root 0 Dec 15 04:08 cron_stderr.log
-rwx------ 1 root root 0 Dec 15 04:08 cron_stdout.log
-rwx------ 1 root root 613 Dec 15 04:08 jupyterhub-upgrade-db_stderr.log
-rwx------ 1 root root 26 Dec 15 04:08 jupyterhub-upgrade-db_stdout.log
-rwx------ 1 root root 77 Dec 15 04:08 jupyterhub_stderr.log
-rwx------ 1 root root 3554 Dec 15 08:03 jupyterhub_stdout.log
-rwx------ 1 root root 14333 Dec 15 08:04 mosquitto_stderr.log
-rwx------ 1 root root 0 Dec 15 04:08 mosquitto_stdout.log
-rwx------ 1 root root 0 Dec 15 04:08 sshd_stderr.log
-rwx------ 1 root root 0 Dec 15 04:08 sshd_stdout.log
-rwx------ 1 root root 0 Dec 15 04:08 xvfb_stderr.log
-rwx------ 1 root root 0 Dec 15 04:08 xvfb_stdout.log
root@5fc5db8bc5cf:/appdata/logs#
BioProxy
docker exec -it bioproxy /bin/bash
root@6c1fca69acf1:/# ls
bin checkstatus.sh etc memcached postgresql-entrypoint.sh run sys
bitnami dev home mnt prepare_letsencrypt_pem.sh sbin tmp
boot docker-entrypoint-initdb.d lib nfs proc server.js usr
certbot_timer.sh docker-entrypoint-preinitdb.d lib64 nfs.sh renew.sh srv var
checkssl.sh docker-entrypoint.sh media opt root super.sh
root@6c1fca69acf1:/#
-------
-------
root@6c1fca69acf1:/var/log/supervisor# ls -l
total 220
-rw-r--r-- 1 root root 45 Dec 15 04:04 certbot_stderr.log
-rw-r--r-- 1 root root 21 Dec 15 04:04 certbot_stdout.log
-rw-r--r-- 1 root root 0 Dec 15 04:04 cron_stderr.log
-rw-r--r-- 1 root root 0 Dec 15 04:04 cron_stdout.log
-rw-r--r-- 1 root root 0 Dec 15 04:04 dataplaneapi_stderr.log
-rw-r--r-- 1 root root 0 Dec 15 04:04 dataplaneapi_stdout.log
-rw-r--r-- 1 root root 997 Dec 15 04:08 haproxy_stderr.log
-rw-r--r-- 1 root root 0 Dec 15 04:04 haproxy_stdout.log
-rw-r--r-- 1 root root 0 Dec 15 04:04 memcached_stderr.log
-rw-r--r-- 1 root root 0 Dec 15 04:04 memcached_stdout.log
-rw-r--r-- 1 root root 0 Dec 15 04:04 nfsserver_stderr.log
-rw-r--r-- 1 root root 170850 Dec 15 08:08 nfsserver_stdout.log
-rw-r--r-- 1 root root 0 Dec 15 04:04 nginx_stderr.log
-rw-r--r-- 1 root root 0 Dec 15 04:04 nginx_stdout.log
-rw-r--r-- 1 root root 0 Dec 15 04:04 pgentrypoint_stderr.log
-rw-r--r-- 1 root root 19 Dec 15 04:04 pgentrypoint_stdout.log
-rw-r--r-- 1 root root 24450 Dec 15 08:09 postgresql_stderr.log
-rw-r--r-- 1 root root 0 Dec 15 04:04 postgresql_stdout.log
-rw-r--r-- 1 root root 0 Dec 15 04:04 redis_stderr.log
-rw-r--r-- 1 root root 0 Dec 15 04:04 redis_stdout.log
-rw-r--r-- 1 root root 0 Dec 15 04:04 sslapi_stderr.log
-rw-r--r-- 1 root root 81 Dec 15 04:04 sslapi_stdout.log
root@6c1fca69acf1:/var/log/supervisor#
Certificate chain ¶
When ever you would have bundle , pem, p7b three files then you must combine pem, bundle.pem and create certificate chain.
cat test.pem test_bundle.pem > test_chained.pem
now you would have test_chained.pem and test.p7b file.
# openssl x509 -text -noout -in test_chained.pem
above command will show the detail and validation.
BioStudio Supervior services ¶
All supervisior service configured in below location.
/etc/supervisor/supervisord.conf
Download t2d services inside BioColab
# cd /appdata/apps/download
# wget "https://colablocal.bioturing.com/content/t2d_blc_tool"
# wget "https://colablocal.bioturing.com/content/t2d_dsc_tool"
# chmod 700 t2d_blc_tool t2d_dsc_tool
# mv t2d_* ../
#
# supervisorctl restart colabblc:*
# supervisorctl restart colab:*
Restart t2d services inside BioColab
# supervisorctl restart colabblc:*
# supervisorctl restart colab:*
root@b1a226ccc20a:/etc/supervisor# supervisorctl restart colabblc:*
colabblc:colabblc_00: stopped
colabblc:colabblc_00: started
Restart haproxy services
root@507cd2637a97:/home# supervisorctl restart haproxy:*
haproxy:haproxy_00: stopped
haproxy:haproxy_00: started
Installing Nginx Proxy ¶
# sudo apt install nginx
root@ip-172-31-39-123:/biocolab/installation-2.1.11/biocolab# curl -I 127.0.0.1
HTTP/1.1 200 OK
Server: nginx/1.18.0 (Ubuntu)
Date: Mon, 18 Dec 2023 07:09:02 GMT
Content-Type: text/html
Content-Length: 612
Last-Modified: Mon, 18 Dec 2023 07:08:18 GMT
Connection: keep-alive
ETag: "657fefe2-264"
Accept-Ranges: bytes
🔔 NOTE: Unti, we will not post vhost entry with Nginx, You can see browser window was Nginx s running well.
Nginx Configuration ¶
# In case of we are not having SSL
server {
listen 0.0.0.0:80;
server_name <Your Domain>.com www.<Your Domain>.com;
#return 301 https:<Your Domain>.com$request_uri;
ignore_invalid_headers off;
client_max_body_size 0;
client_body_timeout 1d;
proxy_buffering off;
proxy_read_timeout 1d;
proxy_connect_timeout 1d;
proxy_send_timeout 1d;
location / {
proxy_pass http://127.0.0.1:8081;
proxy_http_version 1.1;
proxy_set_header X-Real-IP $remote_addr;
proxy_set_header X-Forwarded-For $proxy_add_x_forwarded_for;
proxy_set_header X-Forwarded-Proto $scheme;
proxy_set_header Host $http_host;
add_header X-Host $host;
proxy_set_header X-Forwarded-Host $http_host;
proxy_set_header Upgrade $http_upgrade;
proxy_set_header Connection 'upgrade';
}
location ~ /\.ht {
deny all;
}
}
server {
listen 0.0.0.0:443;
# ssl http2;
server_name <Your Domain>.com www.<Your Domain>.com;
if ($host = 'www.<Your Domain>.com' ) {
rewrite ^/(.*)$ https://<Your Domain>.com/$1 permanent;
}
#ssl_certificate /etc/ssl/certs/testdomain.pem;
#ssl_certificate_key /etc/pki/tls/private/testdomain.key;
#ssl_session_timeout 1d;
#ssl_session_cache shared:SSL:20m;
#ssl_prefer_server_ciphers on;
#ssl_protocols TLSv1 TLSv1.1 TLSv1.2;
#ssl_ciphers EECDH+AESGCM:EDH+AESGCM:AES256+EECDH:AES256+EDH;
ignore_invalid_headers off;
client_max_body_size 0;
client_body_timeout 1d;
proxy_buffering off;
proxy_read_timeout 1d;
proxy_connect_timeout 1d;
proxy_send_timeout 1d;
location / {
proxy_pass http://127.0.0.1:8081;
proxy_http_version 1.1;
proxy_set_header X-Real-IP $remote_addr;
proxy_set_header X-Forwarded-For $proxy_add_x_forwarded_for;
proxy_set_header X-Forwarded-Proto $scheme;
proxy_set_header Host $http_host;
add_header X-Host $host;
proxy_set_header X-Forwarded-Host $http_host;
proxy_set_header Upgrade $http_upgrade;
proxy_set_header Connection 'upgrade';
}
location ~ /\.ht {
deny all;
}
}
Troubleshoot ¶
Not able to install Docker. ¶
⭕ Reason
Ceriticate issue.
curl: (77) error setting certificate file: /etc/ssl/certs/ca-certificates.crt
⭕ Resolution
# echo insecure >> ~/.curlrc
fstab entry for mount point. ¶
⭕ Reason
To auto mount patition.
⭕ Resolution
UUID="3ad2185e-82dd-40cd-8d43-1a82a780d0be" /biocolab xfs defaults 0 0
Error code 413 ( Not able to upload large file). ¶
⭕ Reason
The HTTP 413 Content Too Large response status code signifies that the request entity surpasses the server-defined limits. This issue pertains to resource limitations.
⭕ Resolution
That issue can be fixed by updating the (Nginx) load balancer by incorporating the following value:
client_max_body_size = 0
By implementing this configuration, users will have the flexibility to upload files without any imposed size restrictions. Setting this value to zero will exempt the client request a body size check, allowing users to upload files of unlimited sizes.
- nginx load balancing standalone : client_max_body_size 0
- nginx k8s ingress : nginx.ingress.kubernetes.io/proxy-body-size 0
in case you have seperate Nginx Load Balancer.
nginx.ingress.kubernetes.io/client_max_body_size: 100m
nginx.ingress.kubernetes.io/proxy-body-size: 100m
nginx.ingress.kubernetes.io/proxy-read-timeout: 3600
nginx.ingress.kubernetes.io/proxy-send-timeout: 3600
--Based on situation -- anotation and values can be vary--
Kubernete ingress:
nginx.ingress.kubernetes.io/client_max_body_size: 5000M
nginx.ingress.kubernetes.io/proxy-body-size: 5000M
nginx.ingress.kubernetes.io/proxy-buffer-size: 16k
nginx.ingress.kubernetes.io/proxy-connect-timeout: "3600"
nginx.ingress.kubernetes.io/proxy-read-timeout: "3600"
nginx.ingress.kubernetes.io/proxy-send-timeout: "3600"
nginx.ingress.kubernetes.io/use-regex: "true"
nginx.ingress.kubernetes.io/rewrite-target /;
nginx.ingress.kubernetes.io/ssl_protocols TLSv1 TLSv1.1 TLSv1.2 TLSv1.3;
nginx.ingress.kubernetes.io/client-header-buffer-size 100k;
Nginx lb:
proxy_buffers 4 256k;
proxy_buffer_size 256k;
proxy_busy_buffers_size 256k;
proxy_connect_timeout 3600s;
proxy_read_timeout 3600s;
proxy_send_timeout 3600s;
client_max_body_size 5000M;
For Nginx LB, the value we updated will not be effective until we restart or reload it.
Restart Nginx in case needed:
1] Check Syntax
sudo nginx -t
2] Restart
sudo nginx -s reload
or
sudo systemctl reload nginx
or
sudo systemctl restart nginx
Ref:
Nginx Proxy Module
Nginx HTTP Module
Nginx Suffix rule
Conda installation error. ¶
⭕ Reason
There are several reason that conda driver failed to installed. It could be due to network, blocking, Speed, Software incompatibility with OS ...
⭕ Resolution
Driver can be intalled manually.
Kindly visit below site and choose driver according to OS.
https://developer.nvidia.com/cuda-downloads
root@biocolab-server:/lalit-test-nvidia-driver# sh cuda_12.2.2_535.104.05_linux.run
===========
= Summary =
===========
Driver: Installed
Toolkit: Installed in /usr/local/cuda-12.2/
Please make sure that
- PATH includes /usr/local/cuda-12.2/bin
- LD_LIBRARY_PATH includes /usr/local/cuda-12.2/lib64, or, add /usr/local/cuda-12.2/lib64 to /etc/ld.so.conf and run ldconfig as root
To uninstall the CUDA Toolkit, run cuda-uninstaller in /usr/local/cuda-12.2/bin
To uninstall the NVIDIA Driver, run nvidia-uninstall
Logfile is /var/log/cuda-installer.log
root@biocolab-server:/lalit-test-nvidia-driver#
=====================================
= Solution with cuda version 11.7.1 =
=====================================
sh cuda_11.7.1_515.65.01_linux.run --no-drm
Conda uninstallation. ¶
Sometime, we need to reinstall cuda tool kits. Please follow URL below to uninstall cuda tool kit.
https://docs.nvidia.com/cuda/cuda-installation-guide-linux/index.html
root@biocolab-server:/usr/local/cuda-12.2/bin# pwd
/usr/local/cuda-12.2/bin
root@biocolab-server:/usr/local/cuda-12.2/bin# ls -lhrt cuda*
-rwxr-xr-x 1 root root 15M Sep 22 05:47 cuda-gdb
-rwxr-xr-x 1 root root 789K Sep 22 05:47 cuda-gdbserver
-rwxr-xr-x 1 root root 6.6M Sep 22 05:49 cudafe++
-rwxr-xr-x 1 root root 1023K Sep 22 05:49 cuda-uninstaller
root@biocolab-server:/usr/local/cuda-12.2/bin#
To uninstall cuda driver. Just run uninstallation script.
cd /usr/local/cuda-12.2/bin
root@biocolab-server:/usr/local/cuda-12.2/bin# ./cuda-uninstaller
Successfully uninstalled

Containers are failed to start automatically. ¶
⭕ Reason
There could be several reasons. It could be port mapping or ports are already in use ...etc
⭕ Resolution
Try to identify exact reason by start container manaully by following commands below.
Check docker status
# systemctl status docker.service
# systemctl status docker.service
# docker info
Check container status
# docker ps -a
Run container manually
# docker start biocolab
# docker start bioproxy
Chcek container log to see if we could see any errors
# docker logs biocolab
# docker logs bioproxy
NOTE: Service should be disable which cause port issue. Like NFS.
✏️ Please contact 📧 [support@bioturing.com] and share your issue.
Not able to launch server once try to go inside WORKSPACE. ¶
⭕ Reason
This issue is related to machine creation is not correct and volume. It could be wrong (Private IP) or wrong DNS.
⭕ Resolution
In order to resovle this issue. Please check and make sure that you configured machine private IP / DNS correctly.
Kindly run the following command to check machine IP.
# docker exec biocolab cat /etc/hosts
Check Machine Private IP configured with your DNS.
Note this IP and cross check with Machine IP creation on Admin dashboard.
http://<Domain name>/dashboard.
Kindly check you must create volume attached with this machine.
NOTE: volume name can be anything but Volume path should be "/home"
Not able to download notebook. ¶
⭕ Reason
There could be several reason behind this issue. Common reason is blocking below sites.
*.bioturing.com
*.anaconda.org
*repo.anaconda.com
Amazon S3
github.com
cdn-eu-west-1.s3.eu-west-1.amazonaws.com (euro -west1)
s3.us-west-2.amazonaws.com/cdn.bioturing.com (us -west2)
cdn.bioturing.com (us -west2)
⭕ Resolution
Kindly allow below sites.
*.bioturing.com
*.anaconda.org
*repo.anaconda.com
Amazon S3
github.com
cdn-eu-west-1.s3.eu-west-1.amazonaws.com (euro -west1)
s3.us-west-2.amazonaws.com/cdn.bioturing.com (us -west2)
cdn.bioturing.com (us -west2)
Workspace / any other element access is not showing graphics clear and images are not showing correctly. ¶
⭕ Reason
This issue is related to redirection from Load Balancer.
⭕ Resolution
Please check redirection rule on Load Balancer.
It should be "/*"
So that load balancer allowed all redirections.
Nginx config for BioStudio in case installing on the same server. ¶
Bioturing ecosystem
server {
listen 0.0.0.0:80;
server_name <test-domain.com> www.<test-domain.com>;
return 301 https://<test-domain.com>$request_uri;
}
server {
listen 0.0.0.0:443 ssl http2;
# listen 0.0.0.0:443;
server_name <test-domain.com> www.<test-domain.com>;
if ($host = 'www.<test-domain.com>' ) {
rewrite ^/(.*)$ https://<test-domain.com>/$1 permanent;
}
ssl_certificate /etc/ssl/certs/<test-domain.com>.pem;
ssl_certificate_key /etc/pki/tls/private/<test-domain.com>.key;
ssl_session_timeout 1d;
ssl_session_cache shared:SSL:20m;
ssl_prefer_server_ciphers on;
ssl_protocols TLSv1 TLSv1.1 TLSv1.2;
ssl_ciphers EECDH+AESGCM:EDH+AESGCM:AES256+EECDH:AES256+EDH;
ignore_invalid_headers off;
client_max_body_size 0;
client_body_timeout 1d;
proxy_buffering off;
proxy_read_timeout 1d;
proxy_connect_timeout 1d;
proxy_send_timeout 1d;
location / {
proxy_pass http://127.0.0.1:3000/;
proxy_intercept_errors on;
error_page 404 /404_not_found;
}
}
BioStudio
server {
listen 0.0.0.0:80;
server_name <testdomain>.com www.<testdomain>.com;
return 301 https://<testdomain>.com$request_uri;
ignore_invalid_headers off;
client_max_body_size 0;
client_body_timeout 1d;
proxy_buffering off;
proxy_read_timeout 1d;
proxy_connect_timeout 1d;
proxy_send_timeout 1d;
location / {
proxy_pass http://127.0.0.1:8081;
proxy_http_version 1.1;
proxy_set_header X-Real-IP $remote_addr;
proxy_set_header X-Forwarded-For $proxy_add_x_forwarded_for;
proxy_set_header X-Forwarded-Proto $scheme;
proxy_set_header Host $http_host;
add_header X-Host $host;
proxy_set_header X-Forwarded-Host $http_host;
proxy_set_header Upgrade $http_upgrade;
proxy_set_header Connection 'upgrade';
}
location ~ /\.ht {
deny all;
}
}
server {
listen 0.0.0.0:443 ssl http2;
server_name <testdomain>.com www.<testdomain>.com;
if ($host = 'www.<testdomain>.com' ) {
rewrite ^/(.*)$ https://<testdomain>.com/$1 permanent;
}
ssl_certificate /etc/ssl/certs/testdomain.pem;
ssl_certificate_key /etc/pki/tls/private/testdomain.key;
ssl_session_timeout 1d;
ssl_session_cache shared:SSL:20m;
ssl_prefer_server_ciphers on;
ssl_protocols TLSv1 TLSv1.1 TLSv1.2;
ssl_ciphers EECDH+AESGCM:EDH+AESGCM:AES256+EECDH:AES256+EDH;
ignore_invalid_headers off;
client_max_body_size 0;
client_body_timeout 1d;
proxy_buffering off;
proxy_read_timeout 1d;
proxy_connect_timeout 1d;
proxy_send_timeout 1d;
location / {
proxy_pass http://127.0.0.1:8081;
proxy_http_version 1.1;
proxy_set_header X-Real-IP $remote_addr;
proxy_set_header X-Forwarded-For $proxy_add_x_forwarded_for;
proxy_set_header X-Forwarded-Proto $scheme;
proxy_set_header Host $http_host;
add_header X-Host $host;
proxy_set_header X-Forwarded-Host $http_host;
proxy_set_header Upgrade $http_upgrade;
proxy_set_header Connection 'upgrade';
}
location ~ /\.ht {
deny all;
}
}
============== OR ==============
server {
listen 0.0.0.0:80;
server_name <testdomain>.com www.<testdomain>.com;
return 301 https://<testdomain>.com$request_uri;
}
server {
listen 0.0.0.0:443 ssl http2;
server_name <testdomain>.com www.<testdomain>.com;
if ($host = 'www.<testdomain>.com' ) {
rewrite ^/(.*)$ https://<testdomain>.com/$1 permanent;
}
ssl_certificate /etc/ssl/certs/testdomain.pem;
ssl_certificate_key /etc/pki/tls/private/testdomain.key;
ssl_session_timeout 1d;
ssl_session_cache shared:SSL:20m;
ssl_prefer_server_ciphers on;
ssl_protocols TLSv1 TLSv1.1 TLSv1.2;
ssl_ciphers EECDH+AESGCM:EDH+AESGCM:AES256+EECDH:AES256+EDH;
ignore_invalid_headers off;
client_max_body_size 0;
client_body_timeout 1d;
proxy_buffering off;
proxy_read_timeout 1d;
proxy_connect_timeout 1d;
proxy_send_timeout 1d;
location / {
proxy_pass http://127.0.0.1:8080;
proxy_http_version 1.1;
proxy_set_header X-Real-IP $remote_addr;
proxy_set_header X-Forwarded-For $proxy_add_x_forwarded_for;
proxy_set_header X-Forwarded-Proto $scheme;
proxy_set_header Host $http_host;
add_header X-Host $host;
proxy_set_header X-Forwarded-Host $http_host;
proxy_set_header Upgrade $http_upgrade;
proxy_set_header Connection 'upgrade';
}
location ~ /\.ht {
deny all;
}
}
============== OR ==============
# In case you have external Load balancers
upstream bioturing_router_http {
ip_hash;
server XX.XX.X.XX:80;
server XX.XX.X.XX:80;
}
server {
listen 443 ssl http2;
server_name biocolab.YOUR-DOMAIN;
ssl_certicate /etc/ssl/certs/YOUR-DOMAINyellow.crt;
ssl_certicate_key /etc/ssl/certs/YOUR-DOMAINyellow.key;
ssl_protocols TLSv1 TLSv1.1 TLSv1.2 TLSv1.3;
location / {
# For reverse proxy to backends
proxy_pass http://bioturing_router_http;
proxy_redirect off;
proxy_set_header Host $host;
proxy_set_header X-Real-IP $remote_addr;
proxy_set_header X-Forwarded-For $proxy_add_x_forwarded_for;
proxy_connect_timeout 3600;
proxy_read_timeout 3600;
proxy_write_timeout 3600;
# For HTTP/HTTPS
client_max_body_size 5000m;
client_header_buffer_size 16k;
large_client_header_buffers 4 32k;
# For websocket
proxy_http_version 1.1;
proxy_set_header Upgrade "websocket";
proxy_set_header Connection 'Upgrade';
proxy_cache_bypass "websocket";
proxy_set_header Connection “keep-alive”;
}
}
# In K8s Nginx Ingress. Please help to make sure that the below annotation is set up.
nginx.ingress.kubernetes.io/rewrite-target /;
nginx.ingress.kubernetes.io/ssl_protocols TLSv1 TLSv1.1 TLSv1.2 TLSv1.3;
nginx.ingress.kubernetes.io/client-header-buffer-size 100k;
Multiple redirection and timeout issue. ¶
⭕ Reason
Ingress setting is not correct on LB.
⭕ Resolution
There is no any specific solution. Issue could be form Load Balancer or would be form Application.
⭕ How to test websocket?
BioStudio has an inbuilt feature to test Websocket.
1] Login to BioStudio Admin dashboard.
2] Select System Setting -- Click to Check BioStudio button
You could see the result below.