Notebooks
Premium
Trends
BioTuring
SpatialData (Marconato, Luca, et al., 2023) is a framework for processing spatial omics data, including
- spatialdata-io: load data from common spatial omics technologies into spatialdata.
- spatialdata-plot: static plotting library for spatialdata.
- napari-spatialdata: napari plugin for interactive exploration and annotation of spatial data.
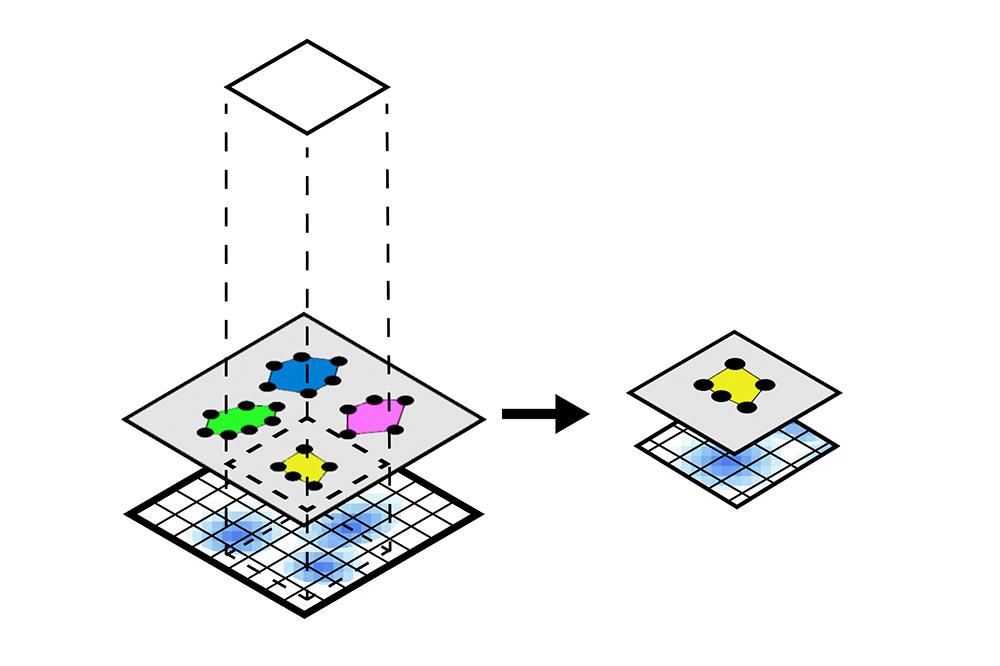
In this notebook, we will illustrate the visualization functions implemented in SpatialData for Visium data. For datasets from other spatial technologies, please check this document. Also, we will use spatial queries to retrieve all the spatial elements and instances that are within a given rectangular window or polygonal shape from an example Visium brain dataset.
The notebook content is inspired from SpatialData's vignette and modified to demonstrate how the tool works on BioTuring's platform.
BioTuring
SpatialData (Marconato, Luca, et al., 2023) is a framework for processing spatial omics data, including
spatialdata-io: load data from common spatial omics technologies into spatialdata.
spatialdata-plot: static plotting library for spatialdata.
napari-spatialdata: napari plugin for interactive exploration and annotation of spatial data.
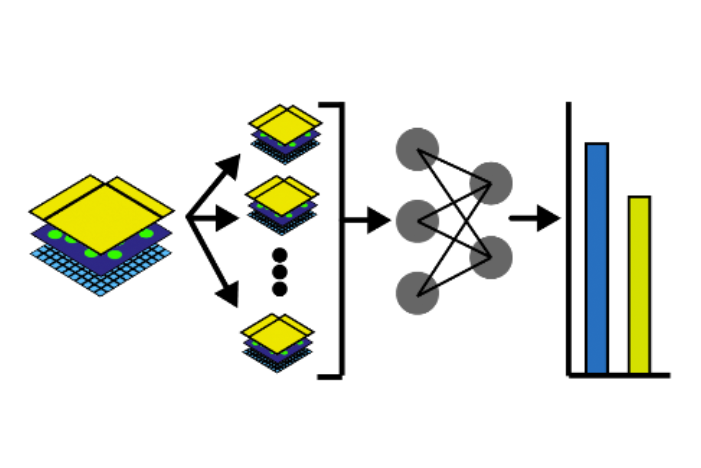
In this notebook, we will illustrate an example to train a Dense Net which predicts cell types Xenium data from an associated H&E image. Particularly, we will access and combine images and annotations across different technologies, where the H&E image from Visium data, and the cell type information from overlapping Xenium data. Also, the two modalities are spatially aligned via an affine transformation.
The notebook content is inspired from SpatialData's vignette and modified to demonstrate how the tool works on BioTuring's platform.